Difference between revisions of "Sentrin-specific protease 1"
(Created page with "{| align="left" | __TOC__ |} {{#invoke:InfoboxforTarget|run|SENP1|[https://www.uniprot.org/uniprot/Q9P0U3 Q9P0U3]|Homo sapiens|Cys603|[http://pfam.xfam.org/family/PF02902...") |
|||
| Line 41: | Line 41: | ||
==Related Pathway== | ==Related Pathway== | ||
| − | : | + | *[https://www.genome.jp/kegg-bin/show_pathway?ko03013 RNA transport] <br/> |
| + | *[https://www.genome.jp/kegg-bin/show_pathway?ko04310 Wnt signaling pathway] <br/> | ||
==Experimental Evidence== | ==Experimental Evidence== | ||
| Line 55: | Line 56: | ||
[[Category:Peptidase C48 family|Peptidase C48 family]] | [[Category:Peptidase C48 family|Peptidase C48 family]] | ||
[[Category:NR3 subfamily|NR3 subfamily]] | [[Category:NR3 subfamily|NR3 subfamily]] | ||
| + | [[Category:RNA transport|RNA transport]] | ||
| + | [[Category:Wnt signaling pathway|Wnt signaling pathway]] | ||
Revision as of 21:15, 19 August 2019
| Basic Information | |
|---|---|
| Short Name | SENP1 |
| UNP ID | Q9P0U3 |
| Organism | Homo sapiens |
| Cys Site | Cys603 |
| Family/Domain | Peptidase C48 family |
| Known Ligand | Ligand list |
| Function Type | Protease |
Summary
Protein Function
Sentrin-specific protease 1 (SENP1) catalyzes maturation SUMO protein (small ubiquitin-related modifier), which causes hydrolysis peptide bond of SUMO is in a conserved sequence Gly-Gly-|-Ala-Thr-Tyr at the C-terminal to be added to the conjugation of other proteins (sumoylation). (From Wikipedia)
Cys Function & Property
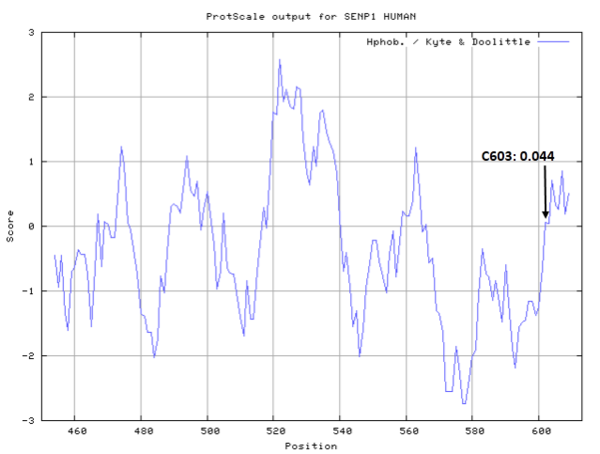
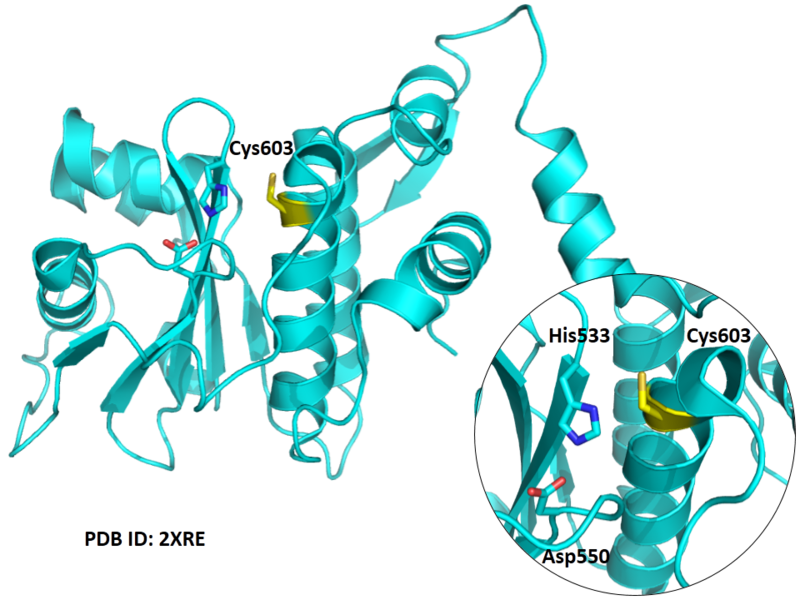
Cys603 is one of the active sites of SENP1.
- Hydrophobic property:
- SASA:
- Cys603: 0.105 A^2
Protein Sequence
MDDIADRMRM DAGEVTLVNH NSVFKTHLLP QTGFPEDQLS LSDQQILSSR
QGHLDRSFTC STRSAAYNPS YYSDNPSSDS FLGSGDLRTF GQSANGQWRN
STPSSSSSLQ KSRNSRSLYL ETRKTSSGLS NSFAGKSNHH CHVSAYEKSF
PIKPVPSPSW SGSCRRSLLS PKKTQRRHVS TAEETVQEEE REIYRQLLQM
VTGKQFTIAK PTTHFPLHLS RCLSSSKNTL KDSLFKNGNS CASQIIGSDT
SSSGSASILT NQEQLSHSVY SLSSYTPDVA FGSKDSGTLH HPHHHHSVPH
QPDNLAASNT QSEGSDSVIL LKVKDSQTPT PSSTFFQAEL WIKELTSVYD
SRARERLRQI EEQKALALQL QNQRLQEREH SVHDSVELHL RVPLEKEIPV
TVVQETQKKG HKLTDSEDEF PEITEEMEKE IKNVFRNGNQ DEVLSEAFRL
TITRKDIQTL NHLNWLNDEI INFYMNMLME RSKEKGLPSV HAFNTFFFTK
LKTAGYQAVK RWTKKVDVFS VDILLVPIHL GVHWCLAVVD FRKKNITYYD
SMGGINNEAC RILLQYLKQE SIDKKRKEFD TNGWQLFSKK SQEIPQQMNG
SDCGMFACKY ADCITKDRPI NFTQQHMPYF RKRMVWEILH RKLL
Structural Information
- Known structure with covalent ligand:
- Unknown
- Protein structure:
Related Pathway
Experimental Evidence
- Homologous Analysis of Sequence, Enzymatic Assay
Reference
- Qiao Z, Wang W, Wang L, et al. Design, synthesis, and biological evaluation of benzodiazepine-based SUMO-specific protease 1 inhibitors[J]. Bioorganic & medicinal chemistry letters, 2011, 21(21): 6389-6392. 21930380
- Ponder E L, Albrow V E, Leader B A, et al. Functional characterization of a SUMO deconjugating protease of Plasmodium falciparum using newly identified small molecule inhibitors[J]. Chemistry & biology, 2011, 18(6): 711-721. 21700207