Difference between revisions of "Glutathione S-transferase P"
(Created page with "{| align="left" | __TOC__ |} {{#invoke:InfoboxforTarget|run|GSTP1-1, GST class-pi|[https://www.uniprot.org/uniprot/P09211 P09211]|Homo sapiens|Cys15, Cys48, Cys102|[http:/...") |
(→Reference) |
||
| Line 60: | Line 60: | ||
# Callan H E, Jenkins R E, Maggs J L, et al. '''Multiple adduction reactions of nitroso sulfamethoxazole with cysteinyl residues of peptides and proteins: implications for hapten formation[J].''' Chemical research in toxicology, 2009, 22(5): 937-948. [https://www.ncbi.nlm.nih.gov/pubmed/?term=19358516 19358516]<br/> | # Callan H E, Jenkins R E, Maggs J L, et al. '''Multiple adduction reactions of nitroso sulfamethoxazole with cysteinyl residues of peptides and proteins: implications for hapten formation[J].''' Chemical research in toxicology, 2009, 22(5): 937-948. [https://www.ncbi.nlm.nih.gov/pubmed/?term=19358516 19358516]<br/> | ||
| − | [[Category: | + | [[Category:Targets]] |
| − | [[Category: | + | [[Category:Homo sapiens]] |
| − | [[Category: | + | [[Category:Metabolic enzyme]] |
| − | [[Category: | + | [[Category:GST superfamily]] |
| − | [[Category: | + | [[Category:Pi family]] |
| − | [[Category: | + | [[Category:Glutathione metabolism]] |
| − | [[Category: | + | [[Category:Metabolism of xenobiotics by cytochrome P450]] |
| − | [[Category: | + | [[Category:Drug metabolism - cytochrome P450]] |
| − | [[Category: | + | [[Category:Drug metabolism - other enzymes]] |
| − | [[Category: | + | [[Category:Metabolic pathways]] |
| − | [[Category: | + | [[Category:Platinum drug resistance]] |
| − | [[Category: | + | [[Category:Pathways in cancer]] |
| − | [[Category: | + | [[Category:Chemical carcinogenesis]] |
| − | [[Category: | + | [[Category:Prostate cancer]] |
| − | [[Category:Hepatocellular carcinoma | + | [[Category:Hepatocellular carcinoma]] |
| − | [[Category: | + | [[Category:Fluid shear stress and atherosclerosis]] |
Revision as of 22:52, 19 August 2019
| Basic Information | |
|---|---|
| Short Name | GSTP1-1, GST class-pi |
| UNP ID | P09211 |
| Organism | Homo sapiens |
| Cys Site | Cys15, Cys48, Cys102 |
| Family/Domain |
Glutathione S-transferase, N-terminal domain, Glutathione S-transferase, C-terminal domain, GST superfamily, Pi family |
| Known Ligand | Ligand list |
| Function Type | Metabolic enzyme |
Summary
Protein Function
Conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles. Regulates negatively CDK5 activity via p25/p35 translocation to prevent neurodegeneration. (From Uniprot)
Glutathione S-transferases (GSTs) are a family of enzymes that play an important role in detoxification by catalyzing the conjugation of many hydrophobic and electrophilic compounds with reduced glutathione. Based on their biochemical, immunologic, and structural properties, the soluble GSTs are categorized into 4 main classes: alpha, mu, pi, and theta.
The glutathione S-transferase pi gene (GSTP1) is a polymorphic gene encoding active, functionally different GSTP1 variant proteins that are thought to function in xenobiotic metabolism and play a role in susceptibility to cancer, and other diseases. (From Wikipedia)
Cys Function & Property
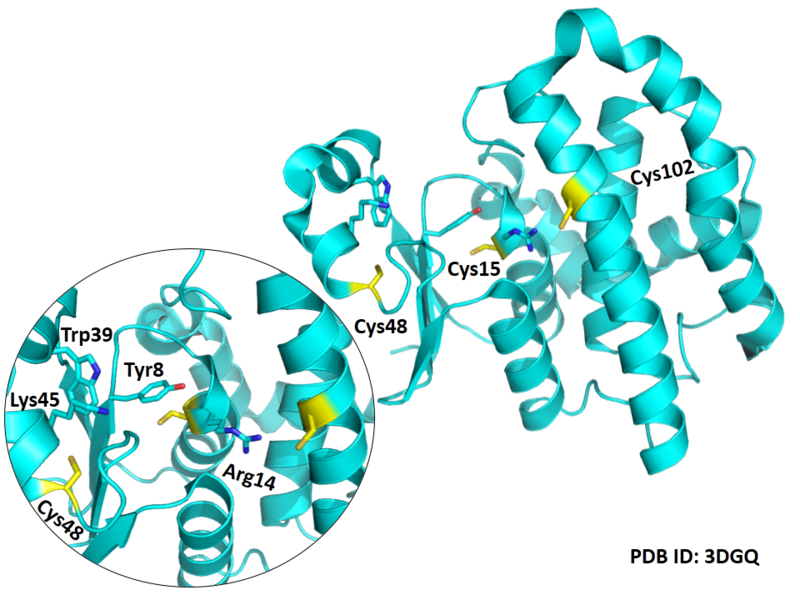
Cys15 and Cys48 are very close to the binding site of Glutathione.
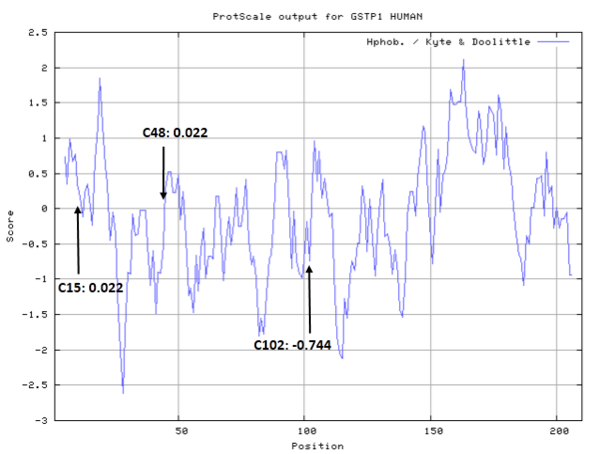
- Hydrophobic property:
- SASA:
- Cys15: 0 A^2
- Cys48: 5.892 A^2
- Cys102: 50.785 A^2
Protein Sequence
MPPYTVVYFP VRGRCAALRM LLADQGQSWK EEVVTVETWQ EGSLKASCLY
GQLPKFQDGD LTLYQSNTIL RHLGRTLGLY GKDQQEAALV DMVNDGVEDL
RCKYISLIYT NYEAGKDDYV KALPGQLKPF ETLLSQNQGG KTFIVGDQIS
FADYNLLDLL LIHEVLAPGC LDAFPLLSAY VGRLSARPKL KAFLASPEYV
NLPINGNGKQ
Structural Information
- Known structures with covalent ligands:
- Unknown
- Protein structure:
Related Pathway
- Glutathione metabolism
- Metabolism of xenobiotics by cytochrome P450
- Drug metabolism - cytochrome P450
- Drug metabolism - other enzymes
- Metabolic pathways
- Platinum drug resistance
- Pathways in cancer
- Chemical carcinogenesis
- Prostate cancer
- Hepatocellular carcinoma
- Fluid shear stress and atherosclerosis
Experimental Evidence
- Cys Modification Assay, LC-MS/MS, Tryptic Digest, Back-titration Experiment, Isothermal Titration Calorimetry, Cys-directed Mutation
Reference
- van Iersel M L P S, Ploemen J P H T M, Bello M L, et al. Interactions of α, β-unsaturated aldehydes and ketones with human glutathione S-transferase P1-1[J]. Chemico-biological interactions, 1997, 108(1-2): 67-78. 9463521
- Boerma J S, Dragovic S, Vermeulen N P E, et al. Mass spectrometric characterization of protein adducts of multiple P450-dependent reactive intermediates of diclofenac to human glutathione-S-transferase P1-1[J]. Chemical research in toxicology, 2012, 25(11): 2532-2541. 22998212
- Meng X, Howarth A, Earnshaw C J, et al. Detection of drug bioactivation in vivo: mechanism of nevirapine–albumin conjugate formation in patients[J]. Chemical research in toxicology, 2013, 26(4): 575-583. 23448204
- Quesada‐Soriano I, Parker L J, Primavera A, et al. Diuretic drug binding to human glutathione transferase P1‐1: potential role of Cys‐101 revealed in the double mutant C47S/Y108V[J]. Journal of Molecular Recognition, 2011, 24(2): 220-234. 20540076
- Callan H E, Jenkins R E, Maggs J L, et al. Multiple adduction reactions of nitroso sulfamethoxazole with cysteinyl residues of peptides and proteins: implications for hapten formation[J]. Chemical research in toxicology, 2009, 22(5): 937-948. 19358516
- Targets
- Homo sapiens
- Metabolic enzyme
- GST superfamily
- Pi family
- Glutathione metabolism
- Metabolism of xenobiotics by cytochrome P450
- Drug metabolism - cytochrome P450
- Drug metabolism - other enzymes
- Metabolic pathways
- Platinum drug resistance
- Pathways in cancer
- Chemical carcinogenesis
- Prostate cancer
- Hepatocellular carcinoma
- Fluid shear stress and atherosclerosis