Difference between revisions of "Ubiquitin-like modifier-activating enzyme 1"
(→Structural Information) |
(→Reference) |
||
| (5 intermediate revisions by the same user not shown) | |||
| Line 53: | Line 53: | ||
*Protein structure: | *Protein structure: | ||
[[File:539-B.png|center|600px]] | [[File:539-B.png|center|600px]] | ||
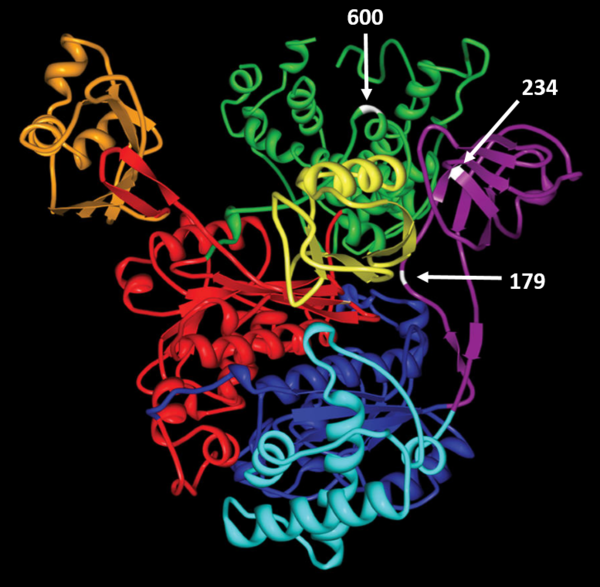
| − | <div align="center">'''Relative locations of adducted residues'''<br/>The active site cysteine Cys600 (white) is located in the SCCH domain.<br/> | + | <div align="center">'''Relative locations of adducted residues'''<br/>The active site cysteine Cys600 (white) is located in the SCCH domain.<br/>Cys179 and Cys234 are located within the FCCH domain of rat E1 (purple). (PMID: 22874009)</div><br/> |
==Related Pathway== | ==Related Pathway== | ||
| Line 65: | Line 65: | ||
# Viquez O M, Caito S W, McDonald W H, et al. '''Electrophilic adduction of ubiquitin activating enzyme E1 by N, N-diethyldithiocarbamate inhibits ubiquitin activation and is accompanied by striatal injury in the rat[J].''' Chemical research in toxicology, 2012, 25(11): 2310-2321. [https://www.ncbi.nlm.nih.gov/pubmed/?term=22874009 22874009]<br/> | # Viquez O M, Caito S W, McDonald W H, et al. '''Electrophilic adduction of ubiquitin activating enzyme E1 by N, N-diethyldithiocarbamate inhibits ubiquitin activation and is accompanied by striatal injury in the rat[J].''' Chemical research in toxicology, 2012, 25(11): 2310-2321. [https://www.ncbi.nlm.nih.gov/pubmed/?term=22874009 22874009]<br/> | ||
| − | [[Category: | + | [[Category:Targets]] |
| − | [[Category: | + | [[Category:Rattus norvegicus]] |
| − | [[Category: | + | [[Category:Ubiquitinase/Deubiquitinase]] |
| − | [[Category: | + | [[Category:Post-translational Modification]] |
| − | [[Category: | + | [[Category:Ubiquitin-activating E1 family]] |
| − | [[Category: | + | [[Category:Ubiquitin mediated proteolysis]] |
| − | [[Category: | + | [[Category:Parkinson disease]] |
Latest revision as of 23:01, 19 August 2019
| Basic Information | |
|---|---|
| Short Name | Uba1, Ube1 |
| UNP ID | Q09472 |
| Organism | Rattus norvegicus |
| Cys Site | Cys179, Cys234 |
| Family/Domain |
ThiF family, Ubiquitin-activating enzyme E1 FCCH domain, Ubiquitin-activating E1 family |
| Known Ligand | Ligand list |
| Function Type |
Ubiquitinase/Deubiquitinase, Post-translational Modification |
Summary
Protein Function
UBA1 participates in ubiquitination and the NEDD8 pathway for protein folding and degradation, among many other biological processes.
This protein encoded by this gene catalyzes the first step in ubiquitin conjugation, or ubiquitination, to mark cellular proteins for degradation. Specifically, UBA1 catalyzes the ATP-dependent adenylation of ubiquitin, thereby forming a thioester bond between the two. It also continues to participate in subsequent steps of ubiquination as a Ub carrier. (From Wikipedia)
ATP + ubiquitin + [E1 ubiquitin-activating enzyme]-L-cysteine = AMP + diphosphate + S-ubiquitinyl-[E1 ubiquitin-activating enzyme]-L-cysteine.
Cys Function & Property
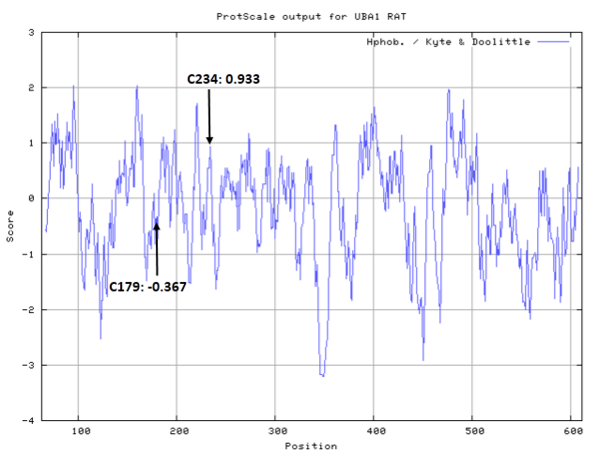
Both Cys234 and Cys179 are located within the first catalytic cysteine domain (FCCH) of E1, a subdomain conserved in human E1 that spans from residues 175–265 and forms one wall of a broad deep groove unique to eukaryotic E1. The rat Cys234 location is identical to that reported for human E1 in vitro, whereas the modified Cys179 on rat E1 represents an additional modification. (PMID: 22874009)
- Hydrophobic property:
- SASA:
- Cys179: Unknown
- Cys234: Unknown
Protein Sequence
MSSSPLSKKR RVSGPDPKPG SNCSSAQSVL SEVSSVPTNG MAKNGSEADI
DESLYSRQLY VLGHEAMKML QTSSVLVSGL RGLGVEIAKN IILGGVKAVT
LHDQGTTQWA DLSSQFYLRE EDIGKNRAEV SQPRLAELNS YVPVTAYTGP
LVEDFLSGFQ VVVLTNSPLE EQLRVGEFCH SRGIKLVVAD TRGLFGQLFC
DFGEEMVLTD SNGEQPLSAM VSMVTKDNPG VVTCLDEARH GFETGDFVSF
SEVQGMVQLN GCQPIEIKVL GPYTFSICDT SNFSDYIRGG IVSQVKVPKK
ISFKSLPASL AEPDFVMTDF AKYSRPAQLH IGFQALHQFC AQHNRPPRPR
NEEDATELVT LAQAVNARSP PAVQQDNVDE DLIRKLAYVA AGDLAPINAF
IGGLAAQEVM KACSGKFMPI MQWLYFDALE CLPEDKEALT EDKCLPRQNR
YDGQVAVFGS DLQEKLGKQK YFLVGAGAIG CELLKNFAMI GLGCGEGGEV
VVTDMDTIEK SNLNRQFLFR PWDVTKLKSD TAAAAVRQMN PYIQVTSHQN
RVGPDTERIY DDDFFQNLDG VANALDNVDA RMYMDRRCVY YRKPLLESGT
LGTKGNVQVV IPFLTESYSS SQDPPEKSIP ICTLKNFPNA IEHTLQWARD
EFEGLFKQPA ENVNQYLTDS KFVERTLRLA GTQPLEVLEA VQRSLVLQRP
QTWGDCVTWA CHHWHTQYCN NIRQLLHNFP PDQLTSSGAP FWSGPKRCPH
PLTFDVNNTL HLDYVMAAAN LFAQTYGLTG SQDRAAVASL LQSVQVPEFT
PKSGVKIHVS DQELQSANAS VDDSRLEELK ATLPSPDKLP GFKMYPIDFE
KDDDSNFHMD FIVAASNLRA ENYDISPADR HKSKLIAGKI IPAIATTTAA
VVGLVCLELY KVVQGHQQLD SYKNGFLNLA LPFFGFSEPL AAPRHQYYNQ
EWTLWDRFEV QGLQPNGEEM TLKQFLDYFK TEHKLEITML SQGVSMLYSF
FMPAAKLKER LDQPMTEIVS RVSKRKLGRH VRALVLELCC NDESGEDVEV
PYVRYTIR
Structural Information
- Known structures with covalent ligands:
- Unknown
- Protein structure:
The active site cysteine Cys600 (white) is located in the SCCH domain.
Cys179 and Cys234 are located within the FCCH domain of rat E1 (purple). (PMID: 22874009)
Related Pathway
Experimental Evidence
- LC/MS/MS, Tryptic Digest, Elastase Digest, Subtilysin Digest
Reference
- Viquez O M, Caito S W, McDonald W H, et al. Electrophilic adduction of ubiquitin activating enzyme E1 by N, N-diethyldithiocarbamate inhibits ubiquitin activation and is accompanied by striatal injury in the rat[J]. Chemical research in toxicology, 2012, 25(11): 2310-2321. 22874009