Difference between revisions of "Caspase-1"
(Created page with "{| align="left" | __TOC__ |} {{#invoke:InfoboxforTarget|run|CASP-1|[https://www.uniprot.org/uniprot/P29466 P29466]|Homo sapiens|Cys285|[http://pfam.xfam.org/family/PF00619...") |
|||
| (3 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
| __TOC__ | | __TOC__ | ||
|} | |} | ||
| − | {{#invoke:InfoboxforTarget|run|CASP-1|[https://www.uniprot.org/uniprot/P29466 P29466]|Homo sapiens|Cys285|[http://pfam.xfam.org/family/ | + | {{#invoke:InfoboxforTarget|run|CASP-1|[https://www.uniprot.org/uniprot/P29466 P29466]|Homo sapiens|Cys285|[http://pfam.xfam.org/family/PF00656 Peptidase C14A family]|[[:Category:Caspase-1|Ligand list]]|Protease}} |
==Summary== | ==Summary== | ||
| Line 31: | Line 31: | ||
==Structural Information== | ==Structural Information== | ||
*Known structure with covalent ligand: <br/> | *Known structure with covalent ligand: <br/> | ||
| − | [https://www.rcsb.org/structure/1ICE 1ICE], [https://www.rcsb.org/structure/3NS7 3NS7], [https://www.rcsb.org/structure/1BMQ 1BMQ], [https://www.rcsb.org/structure/1RWK 1RWK], [https://www.rcsb.org/structure/1RWM 1RWM], [https://www.rcsb.org/structure/1RWN 1RWN],<br/>[https://www.rcsb.org/structure/1RWO 1RWO], [https://www.rcsb.org/structure/1RWP 1RWP], [https://www.rcsb.org/structure/1RWV 1RWV], [https://www.rcsb.org/structure/1RWW 1RWW], [https://www.rcsb.org/structure/1RWX 1RWX], [https://www.rcsb.org/structure/2H4W 2H4W],<br/>[https://www.rcsb.org/structure/2H4Y 2H4Y], [https://www.rcsb.org/structure/2H51 2H51], [https://www.rcsb.org/structure/2H54 2H54], [https://www.rcsb.org/structure/2HBQ 2HBQ], [https://www.rcsb.org/structure/2HBR 2HBR], [https://www.rcsb.org/structure/2HBY 2HBY], | + | [https://www.rcsb.org/structure/1ICE 1ICE], [https://www.rcsb.org/structure/3NS7 3NS7], [https://www.rcsb.org/structure/1BMQ 1BMQ], [https://www.rcsb.org/structure/1RWK 1RWK], [https://www.rcsb.org/structure/1RWM 1RWM], [https://www.rcsb.org/structure/1RWN 1RWN],<br/>[https://www.rcsb.org/structure/1RWO 1RWO], [https://www.rcsb.org/structure/1RWP 1RWP], [https://www.rcsb.org/structure/1RWV 1RWV], [https://www.rcsb.org/structure/1RWW 1RWW], [https://www.rcsb.org/structure/1RWX 1RWX], [https://www.rcsb.org/structure/2H4W 2H4W],<br/>[https://www.rcsb.org/structure/2H4Y 2H4Y], [https://www.rcsb.org/structure/2H51 2H51], [https://www.rcsb.org/structure/2H54 2H54], [https://www.rcsb.org/structure/2HBQ 2HBQ], [https://www.rcsb.org/structure/2HBR 2HBR], [https://www.rcsb.org/structure/2HBY 2HBY], [https://www.rcsb.org/structure/2HBZ 2HBZ] |
*Protein structure: | *Protein structure: | ||
| Line 55: | Line 55: | ||
# Dall E, Brandstetter H. '''Mechanistic and structural studies on legumain explain its zymogenicity, distinct activation pathways, and regulation[J].''' Proceedings of the National Academy of Sciences, 2013, 110(27): 10940-10945. [https://www.ncbi.nlm.nih.gov/pubmed/?term=23776206 23776206]<br/> | # Dall E, Brandstetter H. '''Mechanistic and structural studies on legumain explain its zymogenicity, distinct activation pathways, and regulation[J].''' Proceedings of the National Academy of Sciences, 2013, 110(27): 10940-10945. [https://www.ncbi.nlm.nih.gov/pubmed/?term=23776206 23776206]<br/> | ||
| − | [[Category: | + | [[Category:Targets]] |
| − | [[Category: | + | [[Category:Homo sapiens]] |
| − | [[Category: | + | [[Category:Protease]] |
| − | [[Category: | + | [[Category:Peptidase C14A family]] |
| − | [[Category: | + | [[Category:Necroptosis]] |
| − | [[Category: | + | [[Category:NOD-like receptor signaling pathway]] |
| − | [[Category: | + | [[Category:Cytosolic DNA-sensing pathway]] |
| − | [[Category: | + | [[Category:C-type lectin receptor signaling pathway]] |
| − | [[Category: | + | [[Category:Amyotrophic lateral sclerosis (ALS)]] |
| − | [[Category: | + | [[Category:Salmonella infection]] |
| − | [[Category: | + | [[Category:Pertussis]] |
| − | [[Category: | + | [[Category:Legionellosis]] |
| − | [[Category: | + | [[Category:Yersinia infection]] |
| − | [[Category: | + | [[Category:Influenza A]] |
Latest revision as of 02:16, 4 December 2019
| Basic Information | |
|---|---|
| Short Name | CASP-1 |
| UNP ID | P29466 |
| Organism | Homo sapiens |
| Cys Site | Cys285 |
| Family/Domain | Peptidase C14A family |
| Known Ligand | Ligand list |
| Function Type | Protease |
Summary
Protein Function
Caspase-1 (also known as interleukin 1β-converting enzyme or ICE) is expressed as a procaspase-1 zymogen that is processed into a catalytically competent form through autoproteolysis induced by protein oligomerization in vitro, but may require caspase-5 for efficient activation in vivo. It cleaves IL-1β between an Asp and an Ala, releasing the mature cytokine which is involved in a variety of inflammatory processes. Caspase-1 is important for defense against pathogens. It could cleaves and activates sterol regulatory element binding proteins (SREBPs). It also can promote apoptosis. (From Uniprot, PMID: 16511067)
Cys Function & Property
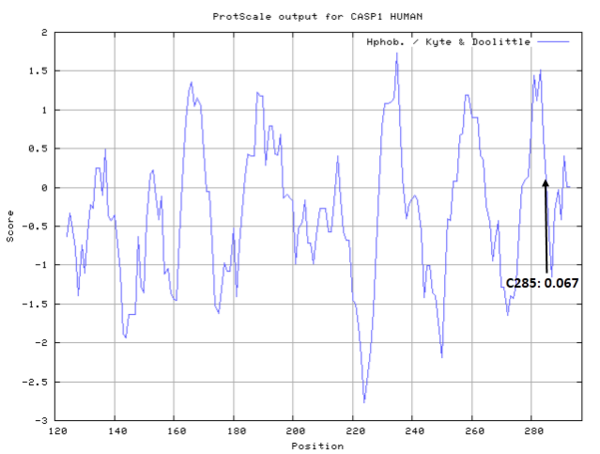
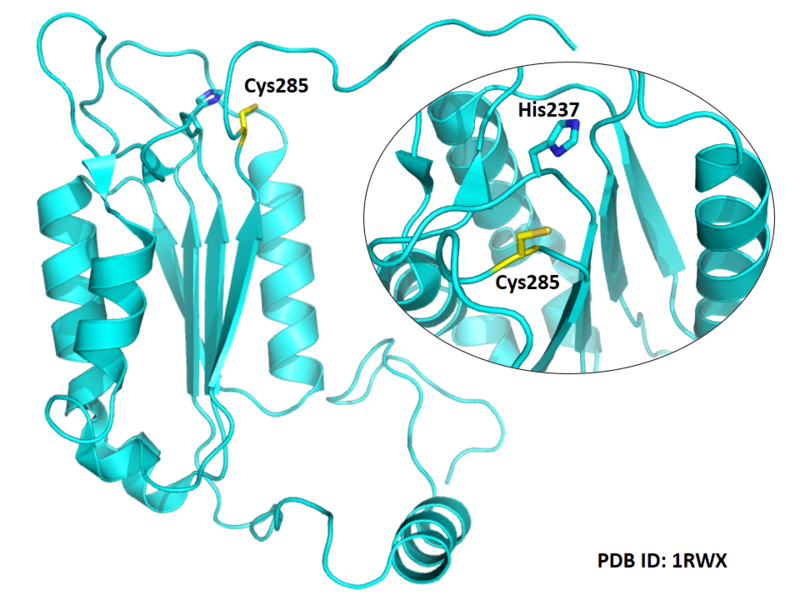
Cys285 is one of the active sites of CASP-1.
- Hydrophobic property:
- SASA:
- Cys285: 11.718 A^2
Protein Sequence
MADKVLKEKR KLFIRSMGEG TINGLLDELL QTRVLNKEEM EKVKRENATV
MDKTRALIDS VIPKGAQACQ ICITYICEED SYLAGTLGLS ADQTSGNYLN
MQDSQGVLSS FPAPQAVQDN PAMPTSSGSE GNVKLCSLEE AQRIWKQKSA
EIYPIMDKSS RTRLALIICN EEFDSIPRRT GAEVDITGMT MLLQNLGYSV
DVKKNLTASD MTTELEAFAH RPEHKTSDST FLVFMSHGIR EGICGKKHSE
QVPDILQLNA IFNMLNTKNC PSLKDKPKVI IIQACRGDSP GVVWFKDSVG
VSGNLSLPTT EEFEDDAIKK AHIEKDFIAF CSSTPDNVSW RHPTMGSVFI
GRLIEHMQEY ACSCDVEEIF RKVRFSFEQP DGRAQMPTTE RVTLTRCFYL
FPGH
Structural Information
- Known structure with covalent ligand:
1ICE, 3NS7, 1BMQ, 1RWK, 1RWM, 1RWN,
1RWO, 1RWP, 1RWV, 1RWW, 1RWX, 2H4W,
2H4Y, 2H51, 2H54, 2HBQ, 2HBR, 2HBY, 2HBZ
- Protein structure:
Related Pathway
- Necroptosis
- NOD-like receptor signaling pathway
- Cytosolic DNA-sensing pathway
- C-type lectin receptor signaling pathway
- Amyotrophic lateral sclerosis (ALS)
- Salmonella infection
- Pertussis
- Legionellosis
- Yersinia infection
- Influenza A
Experimental Evidence
- Crystallography
Reference
- Lee J, Bogyo M. Synthesis and evaluation of aza-peptidyl inhibitors of the lysosomal asparaginyl endopeptidase, legumain[J]. Bioorganic & medicinal chemistry letters, 2012, 22(3): 1340-1343. 22243962
- Dall E, Brandstetter H. Mechanistic and structural studies on legumain explain its zymogenicity, distinct activation pathways, and regulation[J]. Proceedings of the National Academy of Sciences, 2013, 110(27): 10940-10945. 23776206