Difference between revisions of "Ubiquitin-conjugating enzyme E2 L3"
(→Protein Function) |
|||
| Line 8: | Line 8: | ||
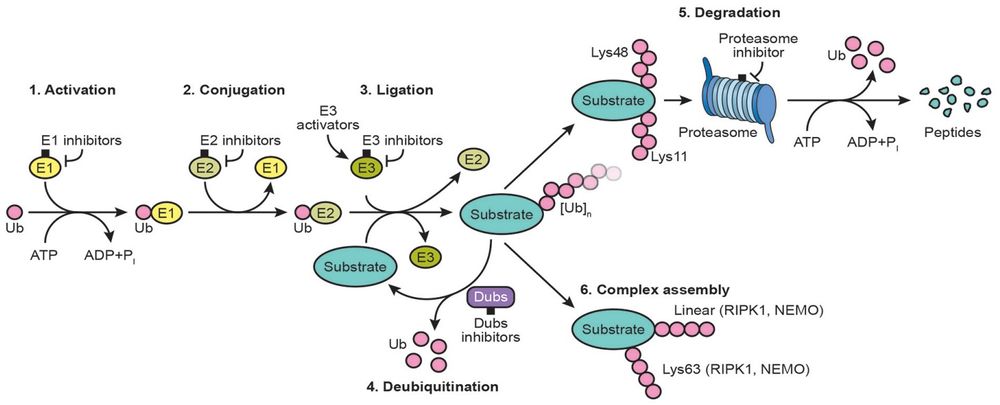
Ubiquitin-conjugating enzyme E2 that specifically acts with HECT-type and RBR family E3 ubiquitin-protein ligases. Does not function with most RING-containing E3 ubiquitin-protein ligases because it lacks intrinsic E3-independent reactivity with lysine: in contrast, it has activity with the RBR family E3 enzymes, such as PRKN and ARIH1, that function like function like RING-HECT hybrids. Accepts ubiquitin from the E1 complex and catalyzes its covalent attachment to other proteins. In vitro catalyzes 'Lys-11'-linked polyubiquitination. Involved in the selective degradation of short-lived and abnormal proteins. Down-regulated during the S-phase it is involved in progression through the cell cycle. Regulates nuclear hormone receptors transcriptional activity. May play a role in myelopoiesis. (From Uniprot)<br/> | Ubiquitin-conjugating enzyme E2 that specifically acts with HECT-type and RBR family E3 ubiquitin-protein ligases. Does not function with most RING-containing E3 ubiquitin-protein ligases because it lacks intrinsic E3-independent reactivity with lysine: in contrast, it has activity with the RBR family E3 enzymes, such as PRKN and ARIH1, that function like function like RING-HECT hybrids. Accepts ubiquitin from the E1 complex and catalyzes its covalent attachment to other proteins. In vitro catalyzes 'Lys-11'-linked polyubiquitination. Involved in the selective degradation of short-lived and abnormal proteins. Down-regulated during the S-phase it is involved in progression through the cell cycle. Regulates nuclear hormone receptors transcriptional activity. May play a role in myelopoiesis. (From Uniprot)<br/> | ||
S-ubiquitinyl-[E1 ubiquitin-activating enzyme]-L-cysteine + [E2 ubiquitin-conjugating enzyme]-L-cysteine = [E1 ubiquitin-activating enzyme]-L-cysteine + S-ubiquitinyl-[E2 ubiquitin-conjugating enzyme]-L-cysteine. <br/> | S-ubiquitinyl-[E1 ubiquitin-activating enzyme]-L-cysteine + [E2 ubiquitin-conjugating enzyme]-L-cysteine = [E1 ubiquitin-activating enzyme]-L-cysteine + S-ubiquitinyl-[E2 ubiquitin-conjugating enzyme]-L-cysteine. <br/> | ||
| + | [[File:571-function-UB.jpg|center|1000px]] | ||
| + | <div align="center">PMID: 27002218</div> | ||
===Cys Function & Property=== | ===Cys Function & Property=== | ||
Revision as of 02:27, 19 August 2019
| Basic Information | |
|---|---|
| Short Name | UbcH7, UBE2L3, Ubiquitin-protein ligase L3 |
| UNP ID | P68036 |
| Organism | Homo sapiens |
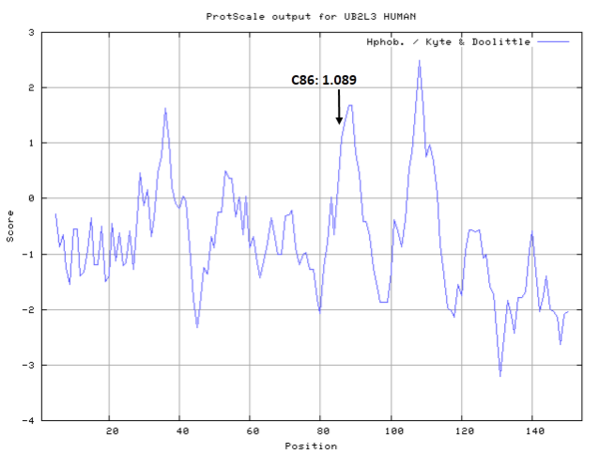
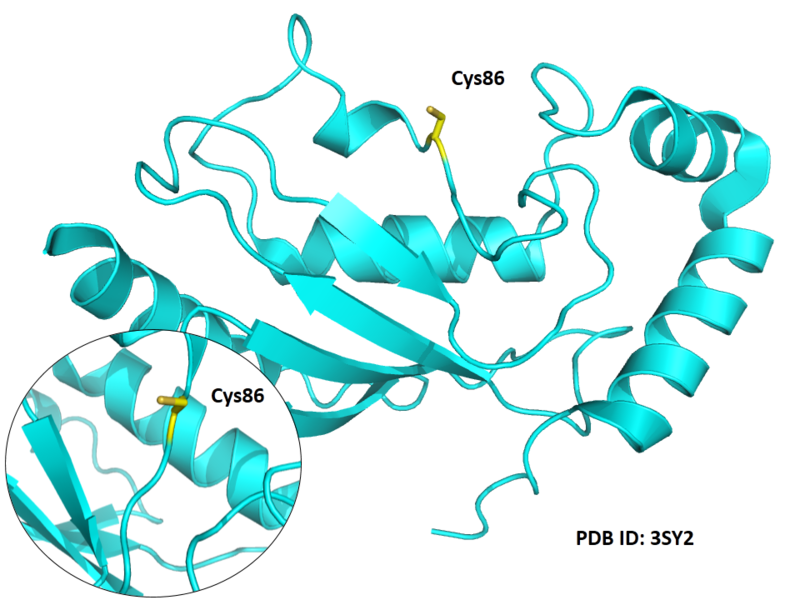
| Cys Site | Cys86 |
| Family/Domain | Ubiquitin-conjugating enzyme family |
| Known Ligand | Ligand list |
| Function Type |
Ubiquitinase/Deubiquitinase, Post-translational Modification |
Summary
Protein Function
Ubiquitin-conjugating enzyme E2 that specifically acts with HECT-type and RBR family E3 ubiquitin-protein ligases. Does not function with most RING-containing E3 ubiquitin-protein ligases because it lacks intrinsic E3-independent reactivity with lysine: in contrast, it has activity with the RBR family E3 enzymes, such as PRKN and ARIH1, that function like function like RING-HECT hybrids. Accepts ubiquitin from the E1 complex and catalyzes its covalent attachment to other proteins. In vitro catalyzes 'Lys-11'-linked polyubiquitination. Involved in the selective degradation of short-lived and abnormal proteins. Down-regulated during the S-phase it is involved in progression through the cell cycle. Regulates nuclear hormone receptors transcriptional activity. May play a role in myelopoiesis. (From Uniprot)
S-ubiquitinyl-[E1 ubiquitin-activating enzyme]-L-cysteine + [E2 ubiquitin-conjugating enzyme]-L-cysteine = [E1 ubiquitin-activating enzyme]-L-cysteine + S-ubiquitinyl-[E2 ubiquitin-conjugating enzyme]-L-cysteine.
Cys Function & Property
Cys87 is the active site of UBE2N, which could form a glycyl thioester intermediate during the catalysis.
- Hydrophobic property:
- SASA:
- Cys87: 32.715 A^2
Protein Sequence
MAASRRLMKE LEEIRKCGMK NFRNIQVDEA NLLTWQGLIV PDNPPYDKGA
FRIEINFPAE YPFKPPKITF KTKIYHPNID EKGQVCLPVI SAENWKPATK
TDQVIQSLIA LVNDPQPEHP LRADLAEEYS KDRKKFCKNA EEFTKKYGEK
RPVD
Structural Information
- Known structures with covalent ligands:
- Unknown
- Protein structure:
Related Pathway
Experimental Evidence
- MALDI-TOF/MS, Tryptic Digest
Reference
- Strickson S, Campbell D G, Emmerich C H, et al. The anti-inflammatory drug BAY 11-7082 suppresses the MyD88-dependent signalling network by targeting the ubiquitin system[J]. Biochemical Journal, 2013, 451(3): 427-437. 23441730