Difference between revisions of "Histone acetyltransferase p300"
(Created page with "{| align="left" | __TOC__ |} {{#invoke:InfoboxforTarget|run|EP300, p300 HAT|[https://www.uniprot.org/uniprot/Q09472 Q09472]|Homo sapiens|Cys1438|[http://pfam.xfam.org/fami...") |
(No difference)
|
Revision as of 20:59, 7 August 2019
| Basic Information | |
|---|---|
| Short Name | EP300, p300 HAT |
| UNP ID | Q09472 |
| Organism | Homo sapiens |
| Cys Site | Cys1438 |
| Family/Domain | Histone acetylation protein |
| Known Ligand | Ligand list |
| Function Type | Post-translational Modification |
Summary
Protein Function
p300 HAT functions as histone acetyltransferase that regulates transcription via chromatin remodeling, and is important in the processes of cell proliferation and differentiation. It mediates cAMP-gene regulation by binding specifically to phosphorylated CREB protein.
p300 HAT contains a bromodomain which is involved in IL6 signaling.
This protein has also been identified as a co-activator of HIF1A (hypoxia-inducible factor 1 alpha), and, thus, plays a role in the stimulation of hypoxia-induced genes such as VEGF. (From Wikipedia)
acetyl-CoA + L-lysyl-[protein] = CoA + H+ + N6-acetyl-L-lysyl-[protein]
Cys Function & Property
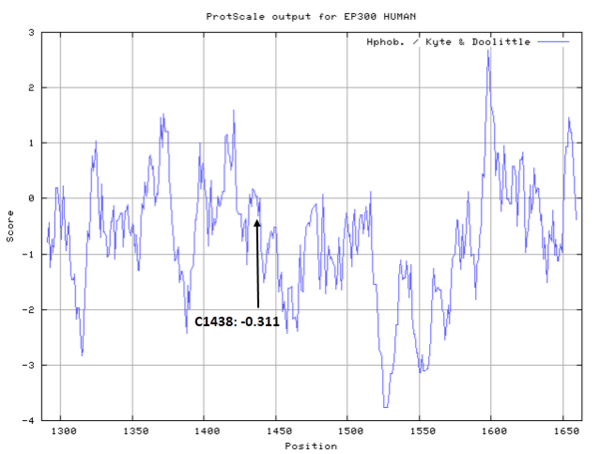
Cys1438 is close to the cofactor Acetyl-CoA binding site in space.
- Hydrophobic property:
- SASA:
- Cys1438: 39.714 A^2
Protein Sequence
MAENVVEPGP PSAKRPKLSS PALSASASDG TDFGSLFDLE HDLPDELINS
TELGLTNGGD INQLQTSLGM VQDAASKHKQ LSELLRSGSS PNLNMGVGGP
GQVMASQAQQ SSPGLGLINS MVKSPMTQAG LTSPNMGMGT SGPNQGPTQS
TGMMNSPVNQ PAMGMNTGMN AGMNPGMLAA GNGQGIMPNQ VMNGSIGAGR
GRQNMQYPNP GMGSAGNLLT EPLQQGSPQM GGQTGLRGPQ PLKMGMMNNP
NPYGSPYTQN PGQQIGASGL GLQIQTKTVL SNNLSPFAMD KKAVPGGGMP
NMGQQPAPQV QQPGLVTPVA QGMGSGAHTA DPEKRKLIQQ QLVLLLHAHK
CQRREQANGE VRQCNLPHCR TMKNVLNHMT HCQSGKSCQV AHCASSRQII
SHWKNCTRHD CPVCLPLKNA GDKRNQQPIL TGAPVGLGNP SSLGVGQQSA
PNLSTVSQID PSSIERAYAA LGLPYQVNQM PTQPQVQAKN QQNQQPGQSP
QGMRPMSNMS ASPMGVNGGV GVQTPSLLSD SMLHSAINSQ NPMMSENASV
PSLGPMPTAA QPSTTGIRKQ WHEDITQDLR NHLVHKLVQA IFPTPDPAAL
KDRRMENLVA YARKVEGDMY ESANNRAEYY HLLAEKIYKI QKELEEKRRT
RLQKQNMLPN AAGMVPVSMN PGPNMGQPQP GMTSNGPLPD PSMIRGSVPN
QMMPRITPQS GLNQFGQMSM AQPPIVPRQT PPLQHHGQLA QPGALNPPMG
YGPRMQQPSN QGQFLPQTQF PSQGMNVTNI PLAPSSGQAP VSQAQMSSSS
CPVNSPIMPP GSQGSHIHCP QLPQPALHQN SPSPVPSRTP TPHHTPPSIG
AQQPPATTIP APVPTPPAMP PGPQSQALHP PPRQTPTPPT TQLPQQVQPS
LPAAPSADQP QQQPRSQQST AASVPTPTAP LLPPQPATPL SQPAVSIEGQ
VSNPPSTSST EVNSQAIAEK QPSQEVKMEA KMEVDQPEPA DTQPEDISES
KVEDCKMEST ETEERSTELK TEIKEEEDQP STSATQSSPA PGQSKKKIFK
PEELRQALMP TLEALYRQDP ESLPFRQPVD PQLLGIPDYF DIVKSPMDLS
TIKRKLDTGQ YQEPWQYVDD IWLMFNNAWL YNRKTSRVYK YCSKLSEVFE
QEIDPVMQSL GYCCGRKLEF SPQTLCCYGK QLCTIPRDAT YYSYQNRYHF
CEKCFNEIQG ESVSLGDDPS QPQTTINKEQ FSKRKNDTLD PELFVECTEC
GRKMHQICVL HHEIIWPAGF VCDGCLKKSA RTRKENKFSA KRLPSTRLGT
FLENRVNDFL RRQNHPESGE VTVRVVHASD KTVEVKPGMK ARFVDSGEMA
ESFPYRTKAL FAFEEIDGVD LCFFGMHVQE YGSDCPPPNQ RRVYISYLDS
VHFFRPKCLR TAVYHEILIG YLEYVKKLGY TTGHIWACPP SEGDDYIFHC
HPPDQKIPKP KRLQEWYKKM LDKAVSERIV HDYKDIFKQA TEDRLTSAKE
LPYFEGDFWP NVLEESIKEL EQEEEERKRE ENTSNESTDV TKGDSKNAKK
KNNKKTSKNK SSLSRGNKKK PGMPNVSNDL SQKLYATMEK HKEVFFVIRL
IAGPAANSLP PIVDPDPLIP CDLMDGRDAF LTLARDKHLE FSSLRRAQWS
TMCMLVELHT QSQDRFVYTC NECKHHVETR WHCTVCEDYD LCITCYNTKN
HDHKMEKLGL GLDDESNNQQ AAATQSPGDS RRLSIQRCIQ SLVHACQCRN
ANCSLPSCQK MKRVVQHTKG CKRKTNGGCP ICKQLIALCC YHAKHCQENK
CPVPFCLNIK QKLRQQQLQH RLQQAQMLRR RMASMQRTGV VGQQQGLPSP
TPATPTTPTG QQPTTPQTPQ PTSQPQPTPP NSMPPYLPRT QAAGPVSQGK
AAGQVTPPTP PQTAQPPLPG PPPAAVEMAM QIQRAAETQR QMAHVQIFQR
PIQHQMPPMT PMAPMGMNPP PMTRGPSGHL EPGMGPTGMQ QQPPWSQGGL
PQPQQLQSGM PRPAMMSVAQ HGQPLNMAPQ PGLGQVGISP LKPGTVSQQA
LQNLLRTLRS PSSPLQQQQV LSILHANPQL LAAFIKQRAA KYANSNPQPI
PGQPGMPQGQ PGLQPPTMPG QQGVHSNPAM QNMNPMQAGV QRAGLPQQQP
QQQLQPPMGG MSPQAQQMNM NHNTMPSQFR DILRRQQMMQ QQQQQGAGPG
IGPGMANHNQ FQQPQGVGYP PQQQQRMQHH MQQMQQGNMG QIGQLPQALG
AEAGASLQAY QQRLLQQQMG SPVQPNPMSP QQHMLPNQAQ SPHLQGQQIP
NSLSNQVRSP QPVPSPRPQS QPPHSSPSPR MQPQPSPHHV SPQTSSPHPG
LVAAQANPME QGHFASPDQN SMLSQLASNP GMANLHGASA TDLGLSTDNS
DLNSNLSQST LDIH
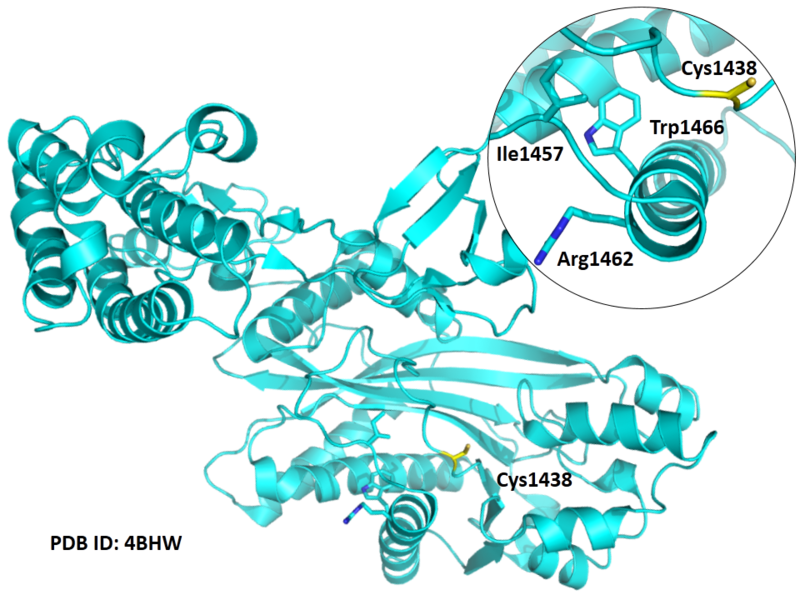
Structural Information
- Known structures with covalent ligands:
- Unknown
- Protein structure:
Related Pathway
- cAMP signaling pathway
- HIF-1 signaling pathway
- FoxO signaling pathway
- Cell cycle
- Wnt signaling pathway
- Notch signaling pathway
- TGF-beta signaling pathway
- Adherens junction
- Jak-STAT signaling pathway
- Long-term potentiation
- Melanogenesis
- Thyroid hormone signaling pathway
- Glucagon signaling pathway
- Huntington disease
- Tuberculosis
- Hepatitis B
- Influenza A
- Human papillomavirus infection
- Human T-cell leukemia virus 1 infection
- Kaposi sarcoma-associated herpesvirus infection
- Pathways in cancer
- Viral carcinogenesis
- MicroRNAs in cancer
- Renal cell carcinoma
- Prostate cancer
Experimental Evidence
- Cys-directed Mutation
Reference
- Pereira S R, Vasconcelos V M, Antunes A. Computational study of the covalent bonding of microcystins to cysteine residues–a reaction involved in the inhibition of the PPP family of protein phosphatases[J]. The FEBS journal, 2013, 280(2): 674-680. 22177231
- MacKintosh R W, Dalby K N, Campbell D G, et al. The cyanobacterial toxin microcystin binds covalently to cysteine-273 on protein phosphatase 1[J]. Febs Letters, 1995, 371(3): 236-240. 7556599
- Hastie C J, Borthwick E B, Morrison L F, et al. Inhibition of several protein phosphatases by a non-covalently interacting microcystin and a novel cyanobacterial peptide, nostocyclin[J]. Biochimica et Biophysica Acta (BBA)-General Subjects, 2005, 1726(2): 187-193. 16046071
- Targets
- Homo sapiens
- Post-translational Modification
- Histone acetylation protein
- CAMP signaling pathway
- HIF-1 signaling pathway
- FoxO signaling pathway
- Cell cycle
- Wnt signaling pathway
- Notch signaling pathway
- TGF-beta signaling pathway
- Adherens junction
- Jak-STAT signaling pathway
- Long-term potentiation
- Melanogenesis
- Thyroid hormone signaling pathway
- Glucagon signaling pathway
- Huntington disease
- Tuberculosis
- Hepatitis B
- Influenza A
- Human papillomavirus infection
- Human T-cell leukemia virus 1 infection
- Kaposi sarcoma-associated herpesvirus infection
- Pathways in cancer
- Viral carcinogenesis
- MicroRNAs in cancer
- Renal cell carcinoma
- Prostate cancer