Difference between revisions of "DNA topoisomerase 2-alpha"
(Created page with "{| align="left" | __TOC__ |} {{#invoke:InfoboxforTarget|run|TOP2A|[https://www.uniprot.org/uniprot/P11388 P11388]|Homo sapiens|Cys170, Cys216, Cys300,<br/>Cys392, Cys405|[...") |
(No difference)
|
Revision as of 16:19, 17 August 2019
| Basic Information | |
|---|---|
| Short Name | TOP2A |
| UNP ID | P11388 |
| Organism | Homo sapiens |
| Cys Site |
Cys170, Cys216, Cys300, Cys392, Cys405 |
| Family/Domain |
DNA gyrase B, Type II topoisomerase family] |
| Known Ligand | Ligand list |
| Function Type | Transcription Related |
Summary
Protein Function
TOP2A is an enzyme that controls and alters the topologic states of DNA during transcription. This nuclear enzyme is involved in processes such as chromosome condensation, chromatid separation, and the relief of torsional stress that occurs during DNA transcription and replication. It catalyzes the transient breaking and rejoining of two strands of duplex DNA which allows the strands to pass through one another, thus altering the topology of DNA. Two forms of this enzyme exist as likely products of a gene duplication event. The gene encoding this form, alpha, is localized to chromosome 17 and the beta gene is localized to chromosome 3. The gene encoding this enzyme functions as the target for several anticancer agents and a variety of mutations in this gene have been associated with the development of drug resistance. Reduced activity of this enzyme may also play a role in ataxia-telangiectasia (From Wikipedia)
Cys Function & Property
TO be added
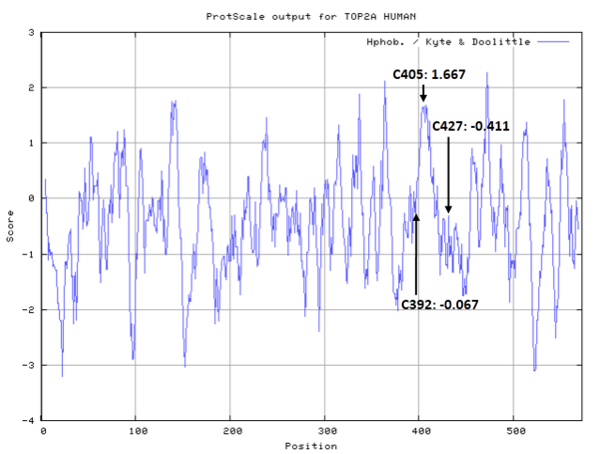
- Hydrophobic property:
- SASA:
- Cys170: 0 A^2
- Cys216: 15.977 A^2
- Cys300: 0 A^2
- Cys392: 0.092 A^2
- Cys405: 6.811 A^2
Protein Sequence
MEVSPLQPVN ENMQVNKIKK NEDAKKRLSV ERIYQKKTQL EHILLRPDTY
IGSVELVTQQ MWVYDEDVGI NYREVTFVPG LYKIFDEILV NAADNKQRDP
KMSCIRVTID PENNLISIWN NGKGIPVVEH KVEKMYVPAL IFGQLLTSSN
YDDDEKKVTG GRNGYGAKLC NIFSTKFTVE TASREYKKMF KQTWMDNMGR
AGEMELKPFN GEDYTCITFQ PDLSKFKMQS LDKDIVALMV RRAYDIAGST
KDVKVFLNGN KLPVKGFRSY VDMYLKDKLD ETGNSLKVIH EQVNHRWEVC
LTMSEKGFQQ ISFVNSIATS KGGRHVDYVA DQIVTKLVDV VKKKNKGGVA
VKAHQVKNHM WIFVNALIEN PTFDSQTKEN MTLQPKSFGS TCQLSEKFIK
AAIGCGIVES ILNWVKFKAQ VQLNKKCSAV KHNRIKGIPK LDDANDAGGR
NSTECTLILT EGDSAKTLAV SGLGVVGRDK YGVFPLRGKI LNVREASHKQ
IMENAEINNI IKIVGLQYKK NYEDEDSLKT LRYGKIMIMT DQDQDGSHIK
GLLINFIHHN WPSLLRHRFL EEFITPIVKV SKNKQEMAFY SLPEFEEWKS
STPNHKKWKV KYYKGLGTST SKEAKEYFAD MKRHRIQFKY SGPEDDAAIS
LAFSKKQIDD RKEWLTNFME DRRQRKLLGL PEDYLYGQTT TYLTYNDFIN
KELILFSNSD NERSIPSMVD GLKPGQRKVL FTCFKRNDKR EVKVAQLAGS
VAEMSSYHHG EMSLMMTIIN LAQNFVGSNN LNLLQPIGQF GTRLHGGKDS
ASPRYIFTML SSLARLLFPP KDDHTLKFLY DDNQRVEPEW YIPIIPMVLI
NGAEGIGTGW SCKIPNFDVR EIVNNIRRLM DGEEPLPMLP SYKNFKGTIE
ELAPNQYVIS GEVAILNSTT IEISELPVRT WTQTYKEQVL EPMLNGTEKT
PPLITDYREY HTDTTVKFVV KMTEEKLAEA ERVGLHKVFK LQTSLTCNSM
VLFDHVGCLK KYDTVLDILR DFFELRLKYY GLRKEWLLGM LGAESAKLNN
QARFILEKID GKIIIENKPK KELIKVLIQR GYDSDPVKAW KEAQQKVPDE
EENEESDNEK ETEKSDSVTD SGPTFNYLLD MPLWYLTKEK KDELCRLRNE
KEQELDTLKR KSPSDLWKED LATFIEELEA VEAKEKQDEQ VGLPGKGGKA
KGKKTQMAEV LPSPRGQRVI PRITIEMKAE AEKKNKKKIK NENTEGSPQE
DGVELEGLKQ RLEKKQKREP GTKTKKQTTL AFKPIKKGKK RNPWSDSESD
RSSDESNFDV PPRETEPRRA ATKTKFTMDL DSDEDFSDFD EKTDDEDFVP
SDASPPKTKT SPKLSNKELK PQKSVVSDLE ADDVKGSVPL SSSPPATHFP
DETEITNPVP KKNVTVKKTA AKSQSSTSTT GAKKRAAPKG TKRDPALNSG
VSQKPDPAKT KNRRKRKPST SDDSDSNFEK IVSKAVTSKK SKGESDDFHM
DFDSAVAPRA KSVRAKKPIK YLEESDEDDL F
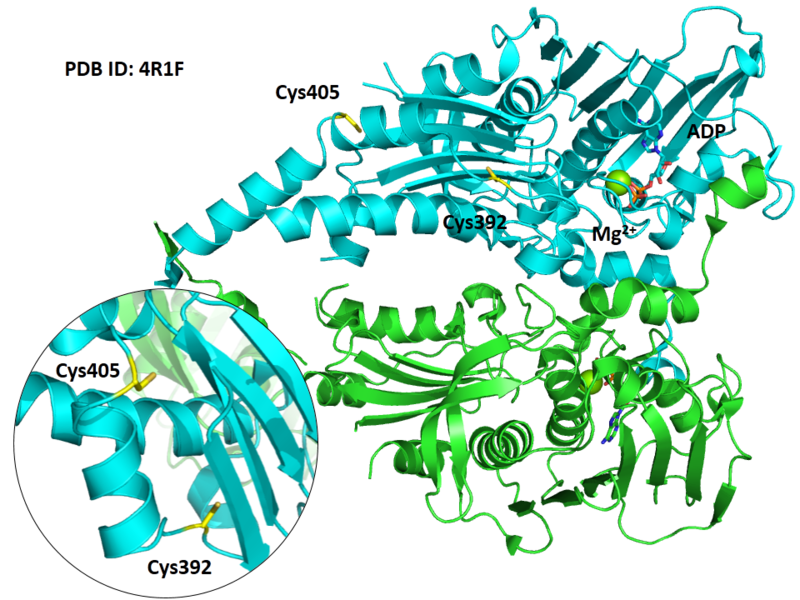
Structural Information
- Known structure with covalent ligand:
- Unknown
- Protein structure:
Related Pathway
Experimental Evidence
- Cys-directed Mutation
Reference
- Wu X, Yalowich J C, Hasinoff B B. Cadmium is a catalytic inhibitor of DNA topoisomerase II[J]. Journal of inorganic biochemistry, 2011, 105(6): 833-838. 21497582