Difference between revisions of "Glutathione S-transferase omega-1"
(Created page with "{| align="left" | __TOC__ |} {{#invoke:InfoboxforTarget|run|GSTO1|[https://www.uniprot.org/uniprot/P78417 P78417]|Homo sapiens|Cys32|[http://pfam.xfam.org/family/PF13417 G...") |
(No difference)
|
Revision as of 05:29, 21 January 2020
| Basic Information | |
|---|---|
| Short Name | GSTO1 |
| UNP ID | P78417 |
| Organism | Homo sapiens |
| Cys Site | Cys32 |
| Family/Domain |
Glutathione S-transferase, GST superfamily, Omega family |
| Known Ligand | Ligand list |
| Function Type | Metabolic enzyme |
Summary
Protein Function
Glutathione S-transferases (GSTs) are a family of phase II detoxification enzymes that conjugate reduced glutathione (GSH) to various electrophilic compounds. GSTs are abundant in cells (making up to 10% of cytosolic protein) and consist of at least seven subfamilies such as alpha, kappa, mu, pi, theta, zeta, and omega, and each subclass has several isoforms, respectively. The omega GSTs (GSTO1 and GSTO2) are a subfamily harboring a cysteine residue in their active site, whereas the other subfamilies have a serine or tyrosine residue. (PMID: 20205472)
GSTO1 is suspected to be involved in certain cancers and neurodegenerative diseases. GSTO1 exhibits glutathione-dependent thiol transferase and dehydroascorbate reductase activities. It also has S-(phenacyl)glutathione reductase and also glutathione S-transferase activity. It could participates in the biotransformation of inorganic arsenic and reduces monomethylarsonic acid (MMA) and dimethylarsonic acid. (From Uniprot)
Cys Function & Property
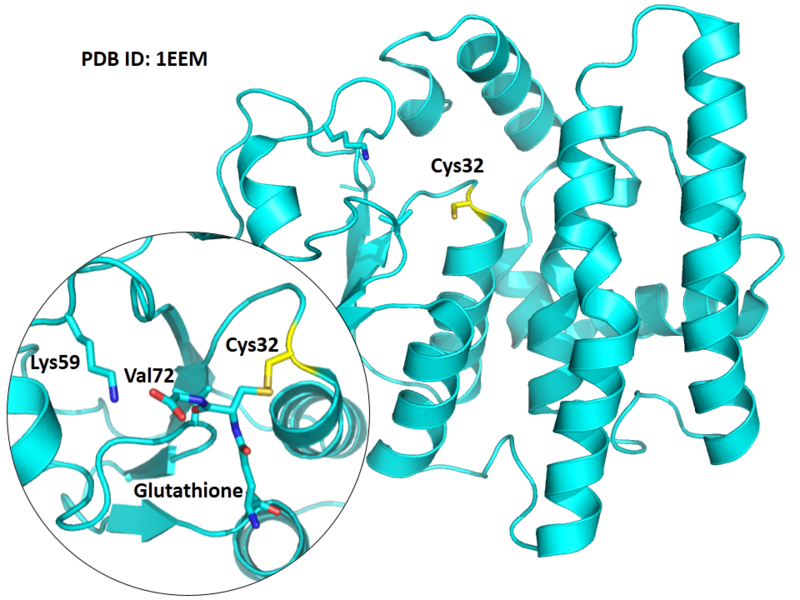
The Cys32 in GSTO1 is the active site, and very close to the Glutathione binding site Lys59 and Val72.
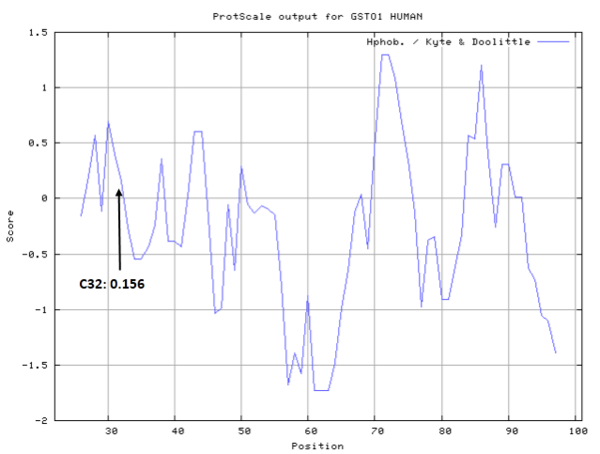
- Hydrophobic property:
- SASA:
- Cys32: Unknown A^2
Protein Sequence
MSGESARSLG KGSAPPGPVP EGSIRIYSMR FCPFAERTRL VLKAKGIRHE
VININLKNKP EWFFKKNPFG LVPVLENSQG QLIYESAITC EYLDEAYPGK
KLLPDDPYEK ACQKMILELF SKVPSLVGSF IRSQNKEDYA GLKEEFRKEF
TKLEEVLTNK KTTFFGGNSI SMIDYLIWPW FERLEAMKLN ECVDHTPKLK
LWMAAMKEDP TVSALLTSEK DWQGFLELYL QNSPEACDYG L
Structural Information
- Known structure with covalent ligand:
- Unknown
- Protein structure:
Related Pathway
- Glutathione metabolism
- Metabolism of xenobiotics by cytochrome P450
- Drug metabolism - cytochrome P450
- Drug metabolism - other enzymes
- Metabolic pathways
- Platinum drug resistance
- Pathways in cancer
- Chemical carcinogenesis
- Hepatocellular carcinoma
- Fluid shear stress and atherosclerosis
Experimental Evidence
- Crystallography, MALDI-TOF-MS/MS, Protein Digest assay
Reference
- Board P G, Coggan M, Chelvanayagam G, et al. Identification, characterization, and crystal structure of the Omega class glutathione transferases[J]. Journal of Biological Chemistry, 2000, 275(32): 24798-24806. 10783391
- Sampayo-Reyes A, Zakharyan R A. Tocopherol esters inhibit human glutathione S-transferase omega[J]. Acta Biochimica Polonica, 2006, 53(3): 547-552. 17019444
- Son J, Lee J J, Lee J S, et al. Isozyme-specific fluorescent inhibitor of glutathione s-transferase omega 1[J]. ACS chemical biology, 2010, 5(5): 449-453. 20205472
- Targets
- Homo sapiens
- Metabolic enzyme
- GST superfamily
- Omega family
- Glutathione metabolism
- Metabolism of xenobiotics by cytochrome P450
- Drug metabolism - cytochrome P450
- Drug metabolism - other enzymes
- Metabolic pathways
- Platinum drug resistance
- Pathways in cancer
- Chemical carcinogenesis
- Hepatocellular carcinoma
- Fluid shear stress and atherosclerosis