Difference between revisions of "Ornithine decarboxylase"
(Created page with "{| align="left" | __TOC__ |} {{#invoke:InfoboxforTarget|run|Odc1|[https://www.uniprot.org/uniprot/P00860 P00860]|Mus musculus|Cys360|[http://pfam.xfam.org/family/PF00278 P...") |
(No difference)
|
Revision as of 03:41, 28 April 2020
| Basic Information | |
|---|---|
| Short Name | Odc1 |
| UNP ID | P00860 |
| Organism | Mus musculus |
| Cys Site | Cys360 |
| Family/Domain | Pyridoxal-dependent decarboxylase, C-terminal sheet domain,Orn/Lys/Arg decarboxylase class-II family |
| Known Ligand | Ligand list |
| Function Type | Metabolic enzyme |
Summary
Protein Function
Eukaryotic ornithine decarboxylase (ODC) is a pyridoxal 5’-phosphate (PLP)-dependent enzyme that catalyzes the initial step in the polyamine biosynthetic pathway, the decarboxylation of L-ornithine to produce putrescine. The polyamines putrescine, spermidine, and spermine are ubiquitous compounds that are essential for cell growth and differentiation in many organisms.
They have demonstrated roles in cell cycle, apoptosis, cancer, embryonic development, immune system functions, and neurochemistry. Overexpression of polyamine biosynthetic enzymes leads to mammalian cell transformation or to growth perturbations in plants, whereas knockout of the polyamine biosynthetic enzymes causes polyamine auxotrophy (e.g. yeast, Trypanosoma brucei, and Leishmania donovani) and is lethal at early embryonic stages in mice. Inhibitors of polyamine biosynthetic enzymes have anti-proliferative effects and have been utilized as anticancer and anti-parasite agents. (PMID: 15190062)
Cys Function & Property
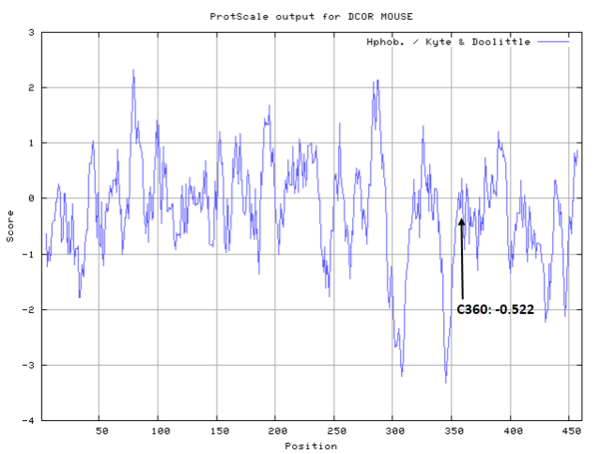
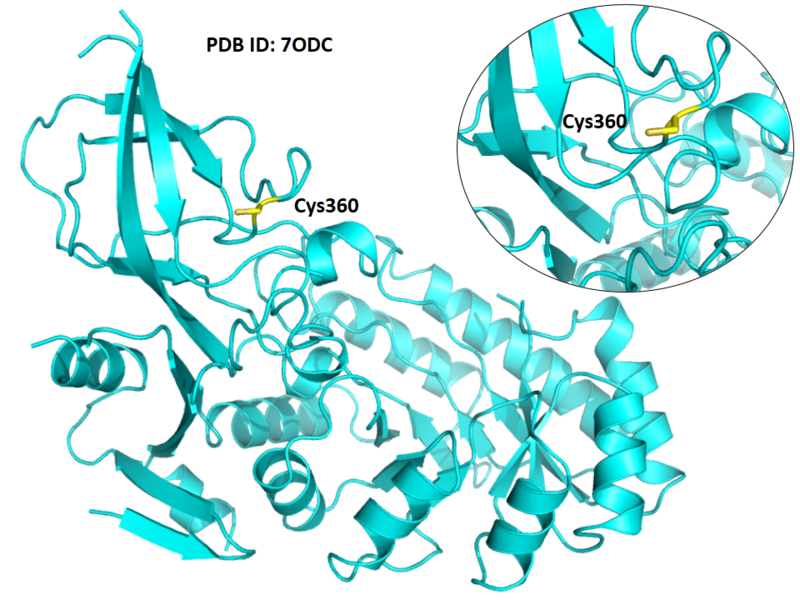
Cys360 is the active site of Odc1 as a proton donor.
- Hydrophobic property:
- SASA:
- Cys360: 121.3 A^2
Protein Sequence
MSSFTKDEFD CHILDEGFTA KDILDQKINE VSSSDDKDAF YVADLGDILK
KHLRWLKALP RVTPFYAVKC NDSRAIVSTL AAIGTGFDCA SKTEIQLVQG
LGVPAERVIY ANPCKQVSQI KYAASNGVQM MTFDSEIELM KVARAHPKAK
LVLRIATDDS KAVCRLSVKF GATLKTSRLL LERAKELNID VIGVSFHVGS
GCTDPETFVQ AVSDARCVFD MATEVGFSMH LLDIGGGFPG SEDTKLKFEE
ITSVINPALD KYFPSDSGVR IIAEPGRYYV ASAFTLAVNI IAKKTVWKEQ
PGSDDEDESN EQTFMYYVND GVYGSFNCIL YDHAHVKALL QKRPKPDEKY
YSSSIWGPTC DGLDRIVERC NLPEMHVGDW MLFENMGAYT VAAASTFNGF
QRPNIYYVMS RPMWQLMKQI QSHGFPPEVE EQDDGTLPMS CAQESGMDRH
PAACASARIN V
Structural Information
- Known structure with covalent ligand:
- Unknown
- Protein structure:
Related Pathway
Experimental Evidence
- Mass Spectrometry, Homologous Analysis of Sequence
Reference
- Shah R, Coleman C S, Mir K, et al. Paramecium bursaria chlorella virus-1 encodes an unusual arginine decarboxylase that is a close homolog of eukaryotic ornithine decarboxylases[J]. Journal of Biological Chemistry, 2004, 279(34): 35760-35767. 15190062
- Poulin R, Lu L, Ackermann B, et al. Mechanism of the irreversible inactivation of mouse ornithine decarboxylase by alpha-difluoromethylornithine. Characterization of sequences at the inhibitor and coenzyme binding sites[J]. Journal of Biological Chemistry, 1992, 267(1): 150-158. 1730582