Difference between revisions of "Protein-glutamine gamma-glutamyltransferase 2"
(Created page with "{| align="left" | __TOC__ |} {{#invoke:InfoboxforTarget|run|TGM2, TGC, TGase H, TGase-2|[https://www.uniprot.org/uniprot/P21980 P21980]|Homo sapiens|Cys277|[http://pfam.xf...") |
(No difference)
|
Latest revision as of 01:53, 30 April 2020
| Basic Information | |
|---|---|
| Short Name | TGM2, TGC, TGase H, TGase-2 |
| UNP ID | P21980 |
| Organism | Homo sapiens |
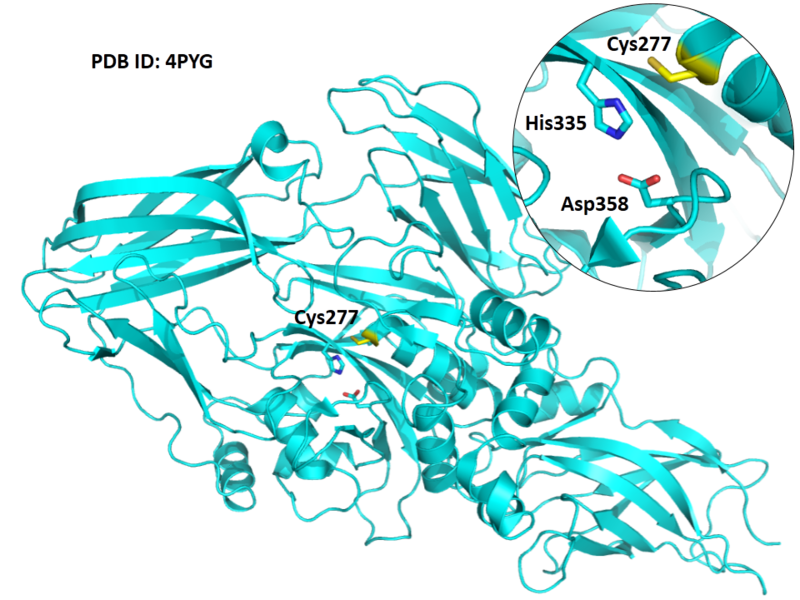
| Cys Site | Cys277 |
| Family/Domain | Transglutaminase-like superfamily |
| Known Ligand | Ligand list |
| Function Type | Metabolic enzyme |
Summary
Protein Function
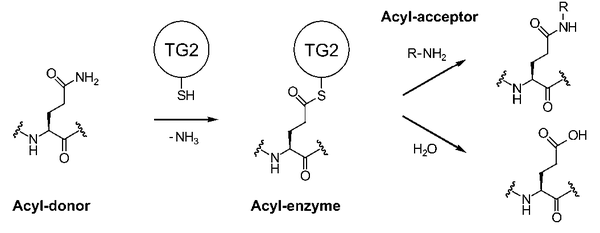
Transglutaminases play important roles in diverse biological functions by selectively crosslinking proteins. They catalyze, in a Ca2+dependent manner, the transamidation of glutamine residues to lysine residues, resulting in proteolytically resistant NƐ(ƴ-glutamyl)lysyl isopeptide bonds.
Transglutaminase 2 (TG2, also known as tissue transglutaminase) s structurally and mechanistically complex, and has both intracellular and extracellular functions. The catalytic mechanism, related to that of cysteine Metabolic enzymes, involves an active site thiol that reacts with a glutamine side chain of a protein or peptide substrate to form a thioester intermediate from which the acyl group is transferred to an amine substrate. In the absence of a suitable amine, water can act as an alternative nucleophile, leading to deamidation of the glutamine residue to glutamate. (PMID: 18092889)
Cys Function & Property
Cys227 is one of the active site of TGM2.
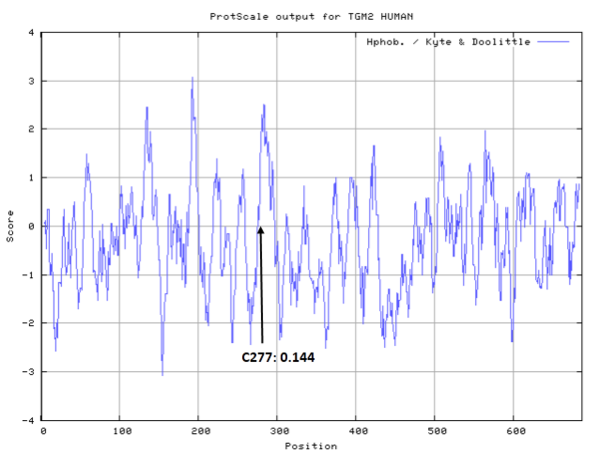
- Hydrophobic property:
- SASA:
- Cys277: 1.107 A^2
Protein Sequence
MAEELVLERC DLELETNGRD HHTADLCREK LVVRRGQPFW LTLHFEGRNY
EASVDSLTFS VVTGPAPSQE AGTKARFPLR DAVEEGDWTA TVVDQQDCTL
SLQLTTPANA PIGLYRLSLE ASTGYQGSSF VLGHFILLFN AWCPADAVYL
DSEEERQEYV LTQQGFIYQG SAKFIKNIPW NFGQFEDGIL DICLILLDVN
PKFLKNAGRD CSRRSSPVYV GRVVSGMVNC NDDQGVLLGR WDNNYGDGVS
PMSWIGSVDI LRRWKNHGCQ RVKYGQCWVF AAVACTVLRC LGIPTRVVTN
YNSAHDQNSN LLIEYFRNEF GEIQGDKSEM IWNFHCWVES WMTRPDLQPG
YEGWQALDPT PQEKSEGTYC CGPVPVRAIK EGDLSTKYDA PFVFAEVNAD
VVDWIQQDDG SVHKSINRSL IVGLKISTKS VGRDEREDIT HTYKYPEGSS
EEREAFTRAN HLNKLAEKEE TGMAMRIRVG QSMNMGSDFD VFAHITNNTA
EEYVCRLLLC ARTVSYNGIL GPECGTKYLL NLNLEPFSEK SVPLCILYEK
YRDCLTESNL IKVRALLVEP VINSYLLAER DLYLENPEIK IRILGEPKQK
RKLVAEVSLQ NPLPVALEGC TFTVEGAGLT EEQKTVEIPD PVEAGEEVKV
RMDLLPLHMG LHKLVVNFES DKLKAVKGFR NVIIGPA
Structural Information
- Known structure with covalent ligand:
- Protein structure:
Related Pathway
Experimental Evidence
- Crystallography
Reference
- Pinkas D M, Strop P, Brunger A T, et al. Transglutaminase 2 undergoes a large conformational change upon activation[J]. PLoS biology, 2007, 5(12). 18092889