Difference between revisions of "Cathepsin S"
(→Reference) |
|||
| Line 2: | Line 2: | ||
| __TOC__ | | __TOC__ | ||
|} | |} | ||
| − | {{#invoke:InfoboxforTarget|run|CTSS|[https://www.uniprot.org/uniprot/P25774 P25774]|Homo sapiens|Cys139|[http://pfam.xfam.org/family/PF00112 Peptidase C1 family]|[[:Category:Cathepsin S|Ligand list]]}} | + | {{#invoke:InfoboxforTarget|run|CTSS|[https://www.uniprot.org/uniprot/P25774 P25774]|Homo sapiens|Cys139|[http://pfam.xfam.org/family/PF00112 Peptidase C1 family]|[[:Category:Cathepsin S|Ligand list]]|Protease}} |
==Summary== | ==Summary== | ||
| Line 53: | Line 53: | ||
[[Category:Targets|Targets]] | [[Category:Targets|Targets]] | ||
[[Category:Homo sapiens|Homo sapiens]] | [[Category:Homo sapiens|Homo sapiens]] | ||
| + | [[Category:Protease|Protease]] | ||
[[Category:Peptidase C1 family|Peptidase C1 family]] | [[Category:Peptidase C1 family|Peptidase C1 family]] | ||
[[Category:Lysosome|Lysosome]] | [[Category:Lysosome|Lysosome]] | ||
Revision as of 20:36, 29 July 2019
| Basic Information | |
|---|---|
| Short Name | CTSS |
| UNP ID | P25774 |
| Organism | Homo sapiens |
| Cys Site | Cys139 |
| Family/Domain | Peptidase C1 family |
| Known Ligand | Ligand list |
| Function Type | Protease |
Summary
Protein Function
Cathepsin S is a thiol protease, which act as key protease responsible for the removal of the invariant chain from MHC class II molecules. The bond-specificity of this proteinase is in part similar to the specificities of cathepsin L and cathepsin N. Similar to cathepsin L, but with much less activity on Z-Phe-Arg-|-NHMec, and more activity on the Z-Val-Val-Arg-|-Xaa compound. (From Uniprot)
Cys Function & Property
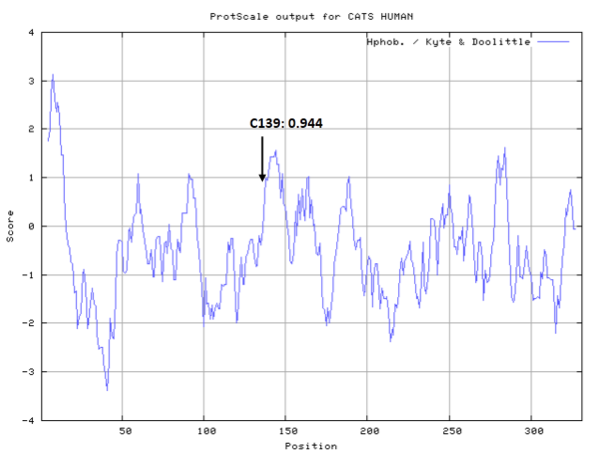
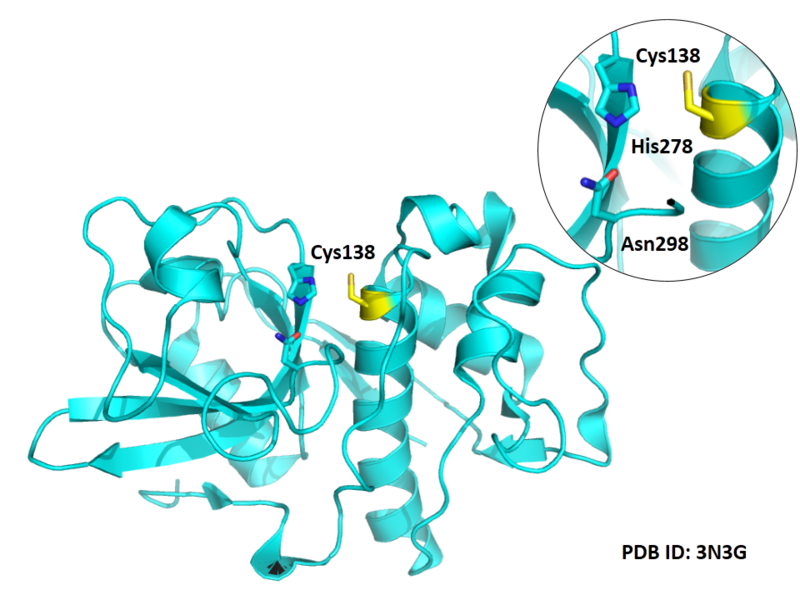
Cys139 is one of the active sites of Cathepsin S, which is very close to His278 and Asn298 in space. These three residues formed a typical catalytic triad motif.
- Hydrophobic property:
- SASA:
- Cys139: 11.884 A^2
Protein Sequence
MKRLVCVLLV CSSAVAQLHK DPTLDHHWHL WKKTYGKQYK EKNEEAVRRL
IWEKNLKFVM LHNLEHSMGM HSYDLGMNHL GDMTSEEVMS LMSSLRVPSQ
WQRNITYKSN PNRILPDSVD WREKGCVTEV KYQGSCGACW AFSAVGALEA
QLKLKTGKLV SLSAQNLVDC STEKYGNKGC NGGFMTTAFQ YIIDNKGIDS
DASYPYKAMD QKCQYDSKYR AATCSKYTEL PYGREDVLKE AVANKGPVSV
GVDARHPSFF LYRSGVYYEP SCTQNVNHGV LVVGYGDLNG KEYWLVKNSW
GHNFGEEGYI RMARNKGNHC GIASFPSYPE I
Structural Information
- Known structures with covalent ligands:
- Protein structure:
Related Pathway
Experimental Evidence
- Crystallography, Cys-directed mutation, Homologous Analysis of Sequence, Enzymatic Assay
Reference
- Pauly T A, Sulea T, Ammirati M, et al. Specificity determinants of human cathepsin S revealed by crystal structures of complexes[J]. Biochemistry, 2003, 42(11): 3203-3213. 12641451
- Cai J, Baugh M, Black D, et al. 6-Phenyl-1H-imidazo [4, 5-c] pyridine-4-carbonitrile as cathepsin S inhibitors[J]. Bioorganic & Medicinal Chemistry Letters, 2010, 20(15): 4350-4354. 20598883
- Inagaki H, Tsuruoka H, Hornsby M, et al. Characterization and optimization of selective, nonpeptidic inhibitors of cathepsin S with an unprecedented binding mode[J]. Journal of Medicinal Chemistry, 2007, 50(11): 2693-2699. 17469812
- Bekkali Y, Thomson D S, Betageri R, et al. Identification of a novel class of succinyl-nitrile-based Cathepsin S inhibitors[J]. Bioorganic & Medicinal Chemistry Letters, 2007, 17(9): 2465-2469. 17379516