Dual specificity phosphatase Cdc25A
| Basic Information | |
|---|---|
| Short Name | CDC25A, M-phase inducer phosphatase 1 |
| UNP ID | P30304 |
| Organism | Homo sapiens |
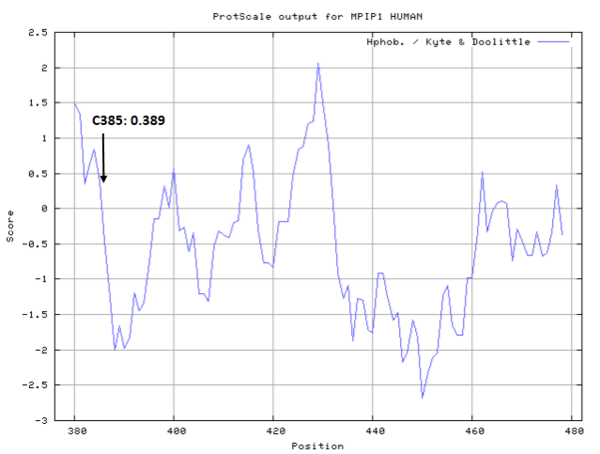
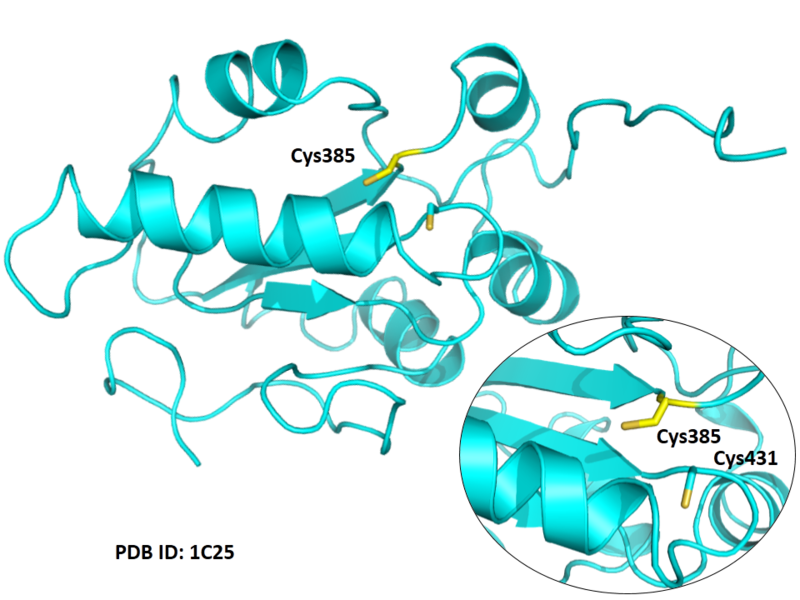
| Cys Site | Cys385 |
| Family/Domain | M-phase inducer (MPI) phosphatase family |
| Known Ligand | Ligand list |
| Function Type |
Phosphatase, Post-translational Modification |
Summary
Protein Function
M-phase inducer phosphatase 1 also known as dual specificity phosphatase Cdc25A is a protein that in humans is encoded by the cell division cycle 25 homolog A (CDC25A) gene. CDC25A is a member of the CDC25 family of dual-specificity phosphatases. Dual-specificity protein phosphatases remove phosphate groups from phosphorylated tyrosine and serine/threonine residues. They represent a subgroup of the tyrosine phosphatase family (as opposed to the serine/threonine phosphatase family). All mammals examined to date have three homologues of the ancestral Cdc25 gene (found e.g. in the fungus species S. pombe , designated Cdc25A, Cdc25B, and Cdc25C. In contrast, some invertebrates harbour 2 (e.g., the Drosophila proteins String and Twine) or four (e.g., C. elegans Cdc-25.1 - Cdc-25.4) homologues. CDC25A is required for progression from G1 to the S phase of the cell cycle, but also plays roles in later cell cycle events. In particular, it is stabilized in metaphase cells and is degraded upon metaphase exit akin to Cyclin B. It is competent to activate the G1/S cyclin-dependent kinases CDK4 and CDK2 by removing inhibitory phosphate groups from adjacent tyrosine and threonine residues; it can also activate Cdc2 (Cdk1), the principal mitotic Cdk.
Cell division cycle 25 molecules (CDC25s) are members of the dual-specificity protein phosphatase (DSP) family of enzymes that dephosphorylate both phospho-serine/threonine and phosphotyrosine residues in the same protein. CDC25s dephosphorylate conserved phospho-tyrosine and phospho-threonine residues on cyclin-dependent kinases (CDKs), thereby activating the kinases and promoting cell cycle progression and transition. In addition to these functions, CDC25s play an important role as checkpoint regulators for handling DNA damage caused by UV light, ionizing irradiation, or chemicals. Therefore, genomic instability is likely to be triggered by misregulation of CDC25s. Mammalian cells express three CDC25 isoforms, CDC25A, CDC25B, and CDC25C. (PMID: 23467652)
CDC25A is required for progression from G1 to the S phase of the cell cycle, but also plays roles in later cell cycle events. In particular, it is stabilized in metaphase cells and is degraded upon metaphase exit akin to Cyclin B. It is competent to activate the G1/S cyclin-dependent kinases CDK4 and CDK2 by removing inhibitory phosphate groups from adjacent tyrosine and threonine residues; it can also activate Cdc2 (Cdk1), the principal mitotic Cdk. (From Wikipedia)
Cys Function & Property
Cys431 is the active site of CDC25B, and Cys385 is very close to this site in space.
- Hydrophobic property:
- SASA:
- Cys385: 0 A^2
Protein Sequence
MELGPEPPHR RRLLFACSPP PASQPVVKAL FGASAAGGLS PVTNLTVTMD
QLQGLGSDYE QPLEVKNNSN LQRMGSSEST DSGFCLDSPG PLDSKENLEN
PMRRIHSLPQ KLLGCSPALK RSHSDSLDHD IFQLIDPDEN KENEAFEFKK
PVRPVSRGCL HSHGLQEGKD LFTQRQNSAP ARMLSSNERD SSEPGNFIPL
FTPQSPVTAT LSDEDDGFVD LLDGENLKNE EETPSCMASL WTAPLVMRTT
NLDNRCKLFD SPSLCSSSTR SVLKRPERSQ EESPPGSTKR RKSMSGASPK
ESTNPEKAHE TLHQSLSLAS SPKGTIENIL DNDPRDLIGD FSKGYLFHTV
AGKHQDLKYI SPEIMASVLN GKFANLIKEF VIIDCRYPYE YEGGHIKGAV
NLHMEEEVED FLLKKPIVPT DGKRVIVVFH CEFSSERGPR MCRYVRERDR
LGNEYPKLHY PELYVLKGGY KEFFMKCQSY CEPPSYRPMH HEDFKEDLKK
FRTKSRTWAG EKSKREMYSR LKKL
Structural Information
- Known structure with covalent ligand:
- Unknown
- Protein structure:
Related Pathway
Experimental Evidence
- MALDI-TOF/MS, Western Blot, Endoproteinase Lys-C Digest, Tryptic Digest
Reference
- Tsuchiya A, Asanuma M, Hirai G, et al. CDC25A-inhibitory RE derivatives bind to pocket adjacent to the catalytic site[J]. Molecular BioSystems, 2013, 9(5): 1026-1034. 23467652