Caspase-2
| Basic Information | |
|---|---|
| Short Name | CASP2 |
| UNP ID | P42575 |
| Organism | Homo sapiens |
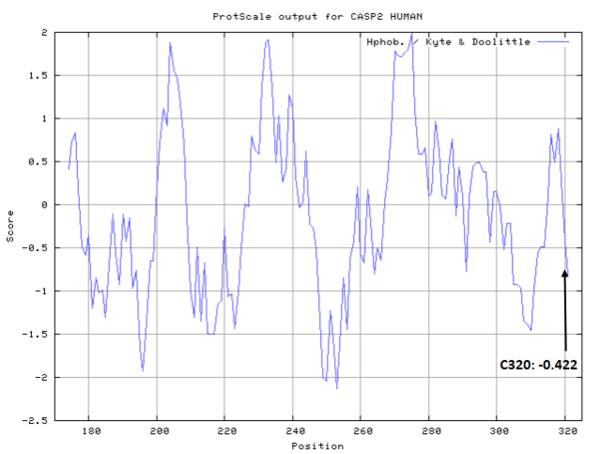
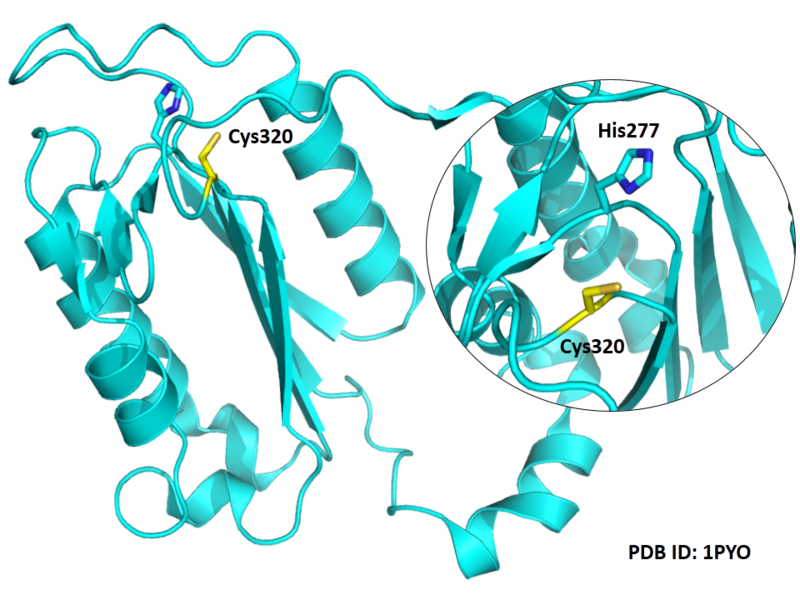
| Cys Site | Cys320 |
| Family/Domain | Peptidase C14A family |
| Known Ligand | Ligand list |
| Function Type | Protease |
Summary
Protein Function
Caspase-2 proteolytically cleaves other proteins. It belongs to a family of cysteine proteases called caspases that cleave proteins only at an amino acid following an aspartic acid residue. Within this family, caspase-2 is part of the Ich-1 subfamily. It is one of the most conserved caspases in different species of animal. Caspase-2 has a similar amino acid sequence to initiator caspases, including caspase 1, caspase-4, caspase-5, and caspase-9. It is produced as a zymogen, which contains a long pro-domain that is similar to that of caspase-9 and contains a protein interaction domain known as a CARD domain. Pro-caspase-2 contains two subunits, p19 and p12. (From Wikipedia)
Caspase-2 was discovered as the first mammalian apoptotic caspase. It is engaged as an initiator in both the extrinsic and the intrinsic pathways of apoptosis. Additionally, Caspase-2 serves in neuronal cells both as the default initiator and the default executioner caspase. (PMID: 12920126)
Cys Function & Property
The catalytic triad in Caspase-3 comprises Cys320, His277 and the backbone carbonyl oxygen atom of Arg219, which points towards the Nϵ atom of His277.
- Hydrophobic property:
- SASA:
- Cys320: 11.39 A^2
Protein Sequence
MAAPSAGSWS TFQHKELMAA DRGRRILGVC GMHPHHQETL KKNRVVLAKQ
LLLSELLEHL LEKDIITLEM RELIQAKVGS FSQNVELLNL LPKRGPQAFD
AFCEALRETK QGHLEDMLLT TLSGLQHVLP PLSCDYDLSL PFPVCESCPL
YKKLRLSTDT VEHSLDNKDG PVCLQVKPCT PEFYQTHFQL AYRLQSRPRG
LALVLSNVHF TGEKELEFRS GGDVDHSTLV TLFKLLGYDV HVLCDQTAQE
MQEKLQNFAQ LPAHRVTDSC IVALLSHGVE GAIYGVDGKL LQLQEVFQLF
DNANCPSLQN KPKMFFIQAC RGDETDRGVD QQDGKNHAGS PGCEESDAGK
EKLPKMRLPT RSDMICGYAC LKGTAAMRNT KRGSWYIEAL AQVFSERACD
MHVADMLVKV NALIKDREGY APGTEFHRCK EMSEYCSTLC RHLYLFPGHP
PT
Structural Information
- Known structure with covalent ligand:
- Protein structure:
Related Pathway
Experimental Evidence
- Crystallography
Reference
- Schweizer A, Briand C, Grütter M G. Crystal structure of caspase-2, apical initiator of the intrinsic apoptotic pathway[J]. Journal of Biological Chemistry, 2003, 278(43): 42441-42447. 12920126