Tubulin beta chain (Homo sapiens)
| Basic Information | |
|---|---|
| Short Name | TUBB |
| UNP ID | P07437 |
| Organism | Homo sapiens |
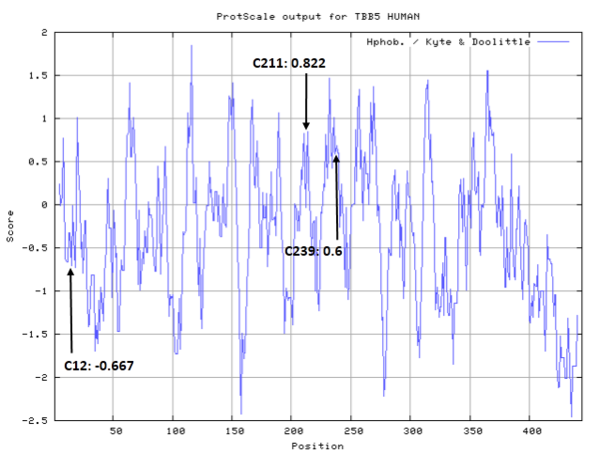
| Cys Site | Cys12, Cys211, Cys239 |
| Family/Domain | Tubulin family |
| Known Ligand | Ligand list |
| Function Type | Structural protein |
Summary
Protein Function
Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain. (From Uniprot)
Tubulin in molecular biology can refer either to the tubulin protein superfamily of globular proteins, or one of the member proteins of that superfamily. The tubulin superfamily contains six families of tubulins (alpha-, beta-, gamma-, delta-, epsilon and zeta-tubulins). Tubulin is also used to specifically refer to α-tubulin and β-tubulin, the proteins that make up microtubules in eukaryotic cells. Each has a molecular weight of approximately 50,000 Daltons. Tubulin was long thought to be specific to eukaryotes. Recently, however, the prokaryotic cell division protein FtsZ was shown to be related to tubulin.
The Tubulin/FtsZ family, GTPase domain is an evolutionary conserved protein domain. This GTPase protein domain is found in all tubulin chains, as well as the bacterial FtsZ family of proteins. These proteins are involved in polymer formation. Tubulin is the major component of microtubules, while FtsZ is the polymer-forming protein of bacterial cell division that forms part of a ring in the middle of the dividing cell that is required for constriction of the cell membrane and cell envelope to yield two daughter cells. FtsZ can polymerise into tubes, sheets, and rings in vitro, and is ubiquitous in bacteria and archaea.
Microtubules are assembled from dimers of α- and β-tubulin. These subunits are slightly acidic with an isoelectric point between 5.2 and 5.8. To form microtubules, the dimers of α- and β-tubulin bind to GTP and assemble onto the (+) ends of microtubules while in the GTP-bound state. The β-tubulin subunit is exposed on the plus end of the microtubule while the α-tubulin subunit is exposed on the minus end. After the dimer is incorporated into the microtubule, the molecule of GTP bound to the β-tubulin subunit eventually hydrolyzes into GDP through inter-dimer contacts along the microtubule protofilament. Whether the β-tubulin member of the tubulin dimer is bound to GTP or GDP influences the stability of the dimer in the microtubule. Dimers bound to GTP tend to assemble into microtubules, while dimers bound to GDP tend to fall apart; thus, this GTP cycle is essential for the dynamic instability of the microtubule. (From Wiki)
Cys Function & Property
From homology structures, these residues are all very close to the interface.
- Hydrophobic property:
- SASA:
- Unknown
Protein Sequence
MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY
YNEATGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDNF VFGQSGAGNN
WAKGHYTEGA ELVDSVLDVV RKESESCDCL QGFQLTHSLG GGTGSGMGTL
LISKIREEYP DRIMNTFSVM PSPKVSDTVV EPYNATLSVH QLVENTDETY
CIDNEALYDI CFRTLKLTTP TYGDLNHLVS ATMSGVTTCL RFPGQLNADL
RKLAVNMVPF PRLHFFMPGF APLTSRGSQQ YRALTVPELT QQMFDSKNMM
AACDPRHGRY LTVAAIFRGR MSMKEVDEQM LNVQNKNSSY FVEWIPNNVK
TAVCDIPPRG LKMSATFIGN STAIQELFKR ISEQFTAMFR RKAFLHWYTG
EGMDEMEFTE AESNMNDLVS EYQQYQDATA DEQGEFEEEE GEDEA
Structural Information
- Known structure with covalent ligand:
- Unknown
- Protein structure:
- Unknown
Related Pathway
Experimental Evidence
- SDS-PAGE, Cys-directed Mutation
Reference
- Li Y M, Broome J D. Arsenic targets tubulins to induce apoptosis in myeloid leukemia cells[J]. Cancer Research, 1999, 59(4): 776-780. 10029061
- Banerjee R, Pace N J, Brown D R, et al. 1, 3, 5-Triazine as a modular scaffold for covalent inhibitors with streamlined target identification[J]. Journal of the American Chemical Society, 2013, 135(7): 2497-2500. 23379904