Cathepsin B (Homo sapiens)
| Basic Information | |
|---|---|
| Short Name | CSPB, CTSB |
| UNP ID | P07858 |
| Organism | Homo sapiens |
| Cys Site | Cys108 |
| Family/Domain | Peptidase C1 family |
| Known Ligand | Ligand list |
| Function Type | Protease |
Summary
Protein Function
Thiol protease which is believed to participate in intracellular degradation and turnover of proteins. Has also been implicated in tumor invasion and metastasis. (From Uniprot)
Cathepsin B (CatB) is an enzymatic protein belonging to the peptidase (or protease) families. In humans, it is coded by the CTSB gene. The protein encoded by this gene is a lysosomal cysteine protease composed of a dimer of disulfide-linked heavy and light chains, both produced from a single protein precursor. It is a member of the peptidase C1 family. At least five transcript variants encoding the same protein have been found for this gene. (From Wikipedia)
Cys Function & Property
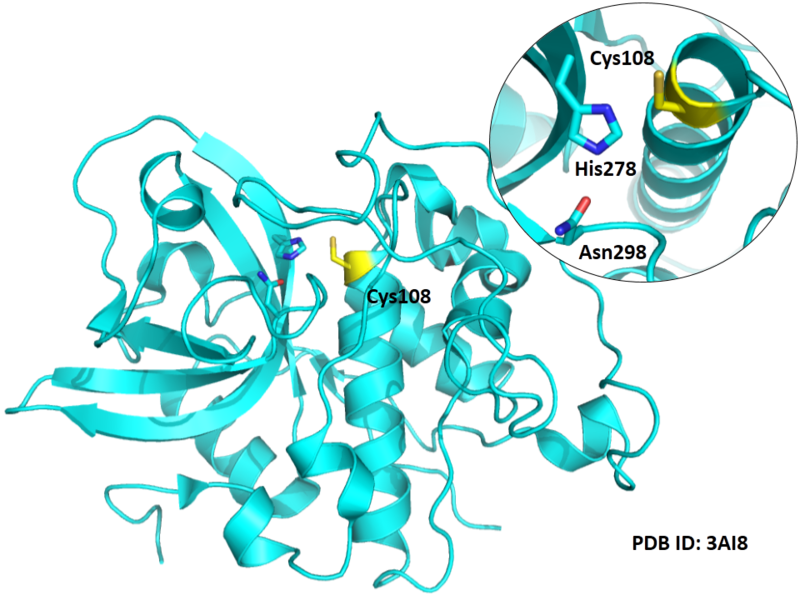
Cys108 is one of the active sites of Cathepsin B, which is very close to His278 and Asn298 in space. These three residues formed a typical catalytic triad motif.
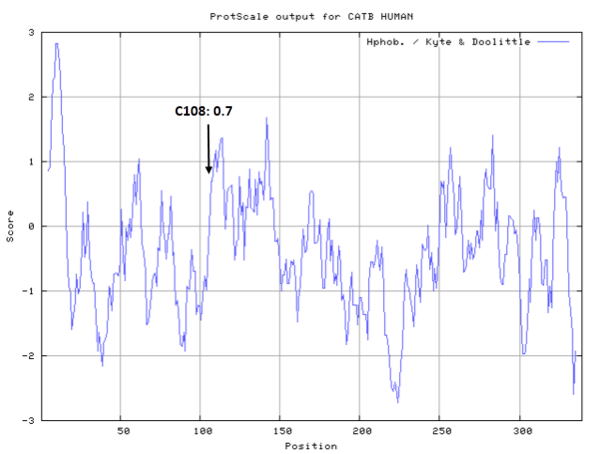
- Hydrophobic property:
- SASA:
- Cys108: 11.819 A^2
Protein Sequence
MWQLWASLCC LLVLANARSR PSFHPLSDEL VNYVNKRNTT WQAGHNFYNV
DMSYLKRLCG TFLGGPKPPQ RVMFTEDLKL PASFDAREQW PQCPTIKEIR
DQGSCGSCWA FGAVEAISDR ICIHTNAHVS VEVSAEDLLT CCGSMCGDGC
NGGYPAEAWN FWTRKGLVSG GLYESHVGCR PYSIPPCEHH VNGSRPPCTG
EGDTPKCSKI CEPGYSPTYK QDKHYGYNSY SVSNSEKDIM AEIYKNGPVE
GAFSVYSDFL LYKSGVYQHV TGEMMGGHAI RILGWGVENG TPYWLVANSW
NTDWGDNGFF KILRGQDHCG IESEVVAGIP RTDQYWEKI
Structural Information
- Known structure with covalent ligand:
- Protein structure:
Related Pathway
- Autophagy - animal
- Lysosome
- Apoptosis
- Antigen processing and presentation
- NOD-like receptor signaling pathway
- Renin secretion
Experimental Evidence
- Crystallography, MALDI-TOF-MS, Homologous Analysis of Sequence, Enzymatic Assay, Isotope Labeling
Reference
- Turk D, Podobnik M, Popovic T, et al. Crystal structure of cathepsin B inhibited with CA030 at 2.0-. ANG. resolution: A basis for the design of specific epoxysuccinyl inhibitors[J]. Biochemistry, 1995, 34(14): 4791-4797. 7718586
- Respondek T, Garner R N, Herroon M K, et al. Light activation of a cysteine protease inhibitor: caging of a peptidomimetic nitrile with RuII(bpy)2[J]. Journal of the American Chemical Society, 2011, 133(43): 17164-17167. 21973207
- Méthot N, Rubin J, Guay D, et al. Inhibition of the activation of multiple serine proteases with a cathepsin C inhibitor requires sustained exposure to prevent pro-enzyme processing[J]. Journal of Biological Chemistry, 2007, 282(29): 20836-20846. 17535802
- Dana D, Davalos A R, De S, et al. Development of cell-active non-peptidyl inhibitors of cysteine cathepsins[J]. Bioorganic & medicinal chemistry, 2013, 21(11): 2975-2987. 23623677
- Greenspan P D, Clark K L, Tommasi R A, et al. Identification of dipeptidyl nitriles as potent and selective inhibitors of cathepsin B through structure-based drug design[J]. Journal of medicinal chemistry, 2001, 44(26): 4524-4534. 11741472
- Wisastra R, Ghizzoni M, Maarsingh H, et al. Isothiazolones; thiol-reactive inhibitors of cysteine protease cathepsin B and histone acetyltransferase PCAF[J]. Organic & biomolecular chemistry, 2011, 9(6): 1817-1822. 21267493
- Falgueyret J P, Black W C, Cromlish W, et al. An activity-based probe for the determination of cysteine cathepsin protease activities in whole cells[J]. Analytical biochemistry, 2004, 335(2): 218-227. 15556560