Cathepsin L1
| Basic Information | |
|---|---|
| Short Name | CTSL, CTSL1 |
| UNP ID | P07711 |
| Organism | Homo sapiens |
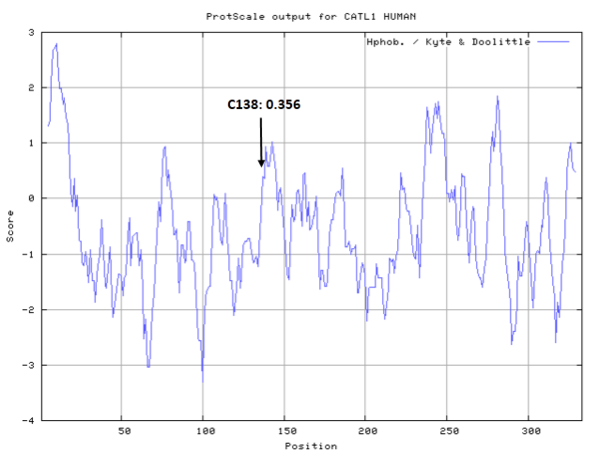
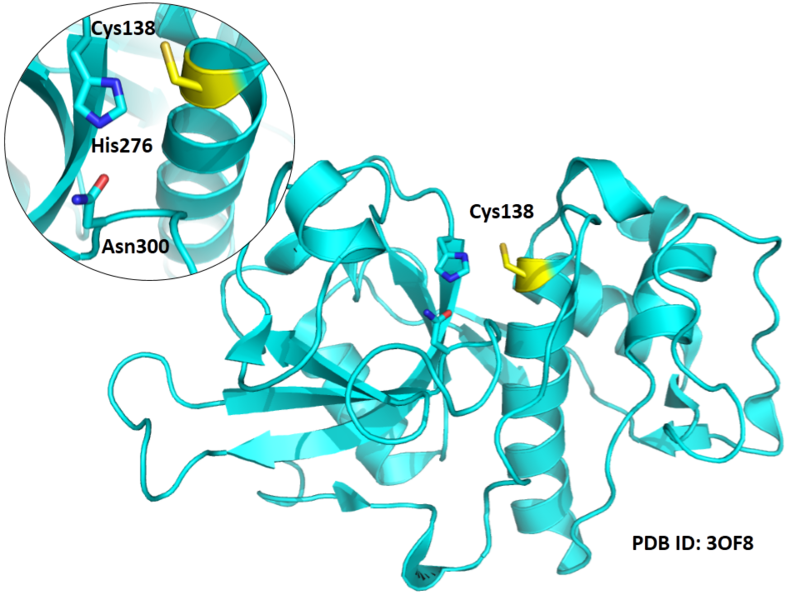
| Cys Site | Cys138 |
| Family/Domain | Peptidase C1 family |
| Known Ligand | Ligand list |
| Function Type | Protease |
Summary
Protein Function
Important for the overall degradation of proteins in lysosomes. (From Uniprot)
Cathepsin L1 is a lysosomal cysteine protease that plays a major role in intracellular protein catabolism. Its substrates include collagen and elastin, as well as alpha-1 protease inhibitor, a major controlling element of neutrophil elastase activity. Cathepsin L1 has been implicated in several pathologic processes, including myofibril necrosis in myopathies and in myocardial ischemia, and in the renal tubular response to proteinuria. This protein, which is a member of the peptidase C1 family, is a dimer composed of disulfide-linked heavy and light chains, both produced from a single protein precursor. At least two transcript variants encoding the same protein have been found for this gene. CTSL1 has been shown to interact with Cystatin A. (From Wikipedia)
Cys Function & Property
Cys138 is one of the active sites of Cathepsin B, which is very close to His276 and Asn300 in space. These three residues formed a typical catalytic triad motif.
- Hydrophobic property:
- SASA:
- Cys138: 11.897 A^2
Protein Sequence
MNPTLILAAF CLGIASATLT FDHSLEAQWT KWKAMHNRLY GMNEEGWRRA
VWEKNMKMIE LHNQEYREGK HSFTMAMNAF GDMTSEEFRQ VMNGFQNRKP
RKGKVFQEPL FYEAPRSVDW REKGYVTPVK NQGQCGSCWA FSATGALEGQ
MFRKTGRLIS LSEQNLVDCS GPQGNEGCNG GLMDYAFQYV QDNGGLDSEE
SYPYEATEES CKYNPKYSVA NDTGFVDIPK QEKALMKAVA TVGPISVAID
AGHESFLFYK EGIYFEPDCS SEDMDHGVLV VGYGFESTES DNNKYWLVKN
SWGEEWGMGG YVKMAKDRRN HCGIASAASY PTV
Structural Information
- Known structure with covalent ligand:
- Protein structure:
Related Pathway
- Autophagy - animal
- Lysosome
- Phagosome
- Apoptosis
- Antigen processing and presentation
- Proteoglycans in cancer
- Rheumatoid arthritis
- Fluid shear stress and atherosclerosis
Experimental Evidence
- Crystallography, Homologous Analysis of Sequence, Enzymatic Assay
Reference
- Torkar A, Lenarčič B, Lah T, et al. Identification of new peptide amides as selective cathepsin L inhibitors: The first step towards selective irreversible inhibitors?[J]. Bioorganic & medicinal chemistry letters, 2013, 23(10): 2968-2973. 23562595
- Respondek T, Garner R N, Herroon M K, et al. Light activation of a cysteine protease inhibitor: caging of a peptidomimetic nitrile with RuII(bpy)2[J]. Journal of the American Chemical Society, 2011, 133(43): 17164-17167. 21973207
Falgueyret J P, Black W C, Cromlish W, et al. An activity-based probe for the determination of cysteine cathepsin protease activities in whole cells[J]. Analytical biochemistry, 2004, 335(2): 218-227. 15556560
- Shenoy, R. T. and J. Sivaraman (2011). "Structural basis for reversible and irreversible inhibition of human cathepsin L by their respective dipeptidyl glyoxal and diazomethylketone inhibitors." J Struct Biol 173(1): 14-19. 20850545
- Hardegger L A, Kuhn B, Spinnler B, et al. Systematic investigation of halogen bonding in protein–ligand interactions[J]. Angewandte Chemie International Edition, 2011, 50(1): 314-318. 21184410
Hardegger L A, Kuhn B, Spinnler B, et al. Halogen bonding at the active sites of human cathepsin L and MEK1 kinase: efficient interactions in different environments[J]. ChemMedChem, 2011, 6(11): 2048-2054. 21898833
- Bethel P A, Gerhardt S, Jones E V, et al. Design of selective cathepsin inhibitors[J]. Bioorganic & medicinal chemistry letters, 2009, 19(16): 4622-4625. 19616430
- Asaad N, Bethel P A, Coulson M D, et al. Dipeptidyl nitrile inhibitors of Cathepsin L[J]. Bioorganic & medicinal chemistry letters, 2009, 19(15): 4280-4283. 19515558