NF-kappa-B p65 (Mus musculus)
| Basic Information | |
|---|---|
| Short Name | NFKB3, Rela, Transcription factor p65 |
| UNP ID | Q04207 |
| Organism | Mus musculus |
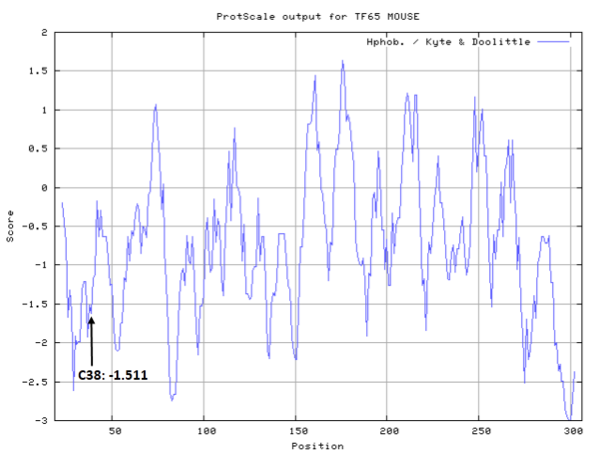
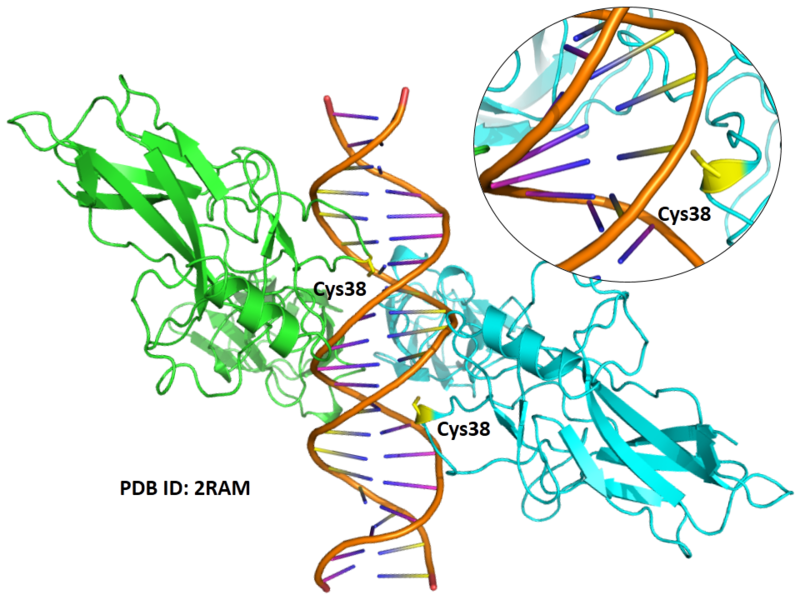
| Cys Site | Cys38 |
| Family/Domain | Rel homology DNA-binding domain |
| Known Ligand | Ligand list |
| Function Type | Transcription factor |
Summary
Protein Function
NF-kappa-B is a pleiotropic transcription factor present in almost all cell types and is the endpoint of a series of signal transduction events that are initiated by a vast array of stimuli related to many biological processes such as inflammation, immunity, differentiation, cell growth, tumorigenesis and apoptosis. NF-kappa-B is a homo- or heterodimeric complex formed by the Rel-like domain-containing proteins RELA/p65, RELB, NFKB1/p105, NFKB1/p50, REL and NFKB2/p52. The heterodimeric RELA-NFKB1 complex appears to be most abundant one. The dimers bind at kappa-B sites in the DNA of their target genes and the individual dimers have distinct preferences for different kappa-B sites that they can bind with distinguishable affinity and specificity. Different dimer combinations act as transcriptional activators or repressors, respectively. The NF-kappa-B heterodimeric RELA-NFKB1 and RELA-REL complexes, for instance, function as transcriptional activators. NF-kappa-B is controlled by various mechanisms of post-translational modification and subcellular compartmentalization as well as by interactions with other cofactors or corepressors. NF-kappa-B complexes are held in the cytoplasm in an inactive state complexed with members of the NF-kappa-B inhibitor (I-kappa-B) family. In a conventional activation pathway, I-kappa-B is phosphorylated by I-kappa-B kinases (IKKs) in response to different activators, subsequently degraded thus liberating the active NF-kappa-B complex which translocates to the nucleus. The inhibitory effect of I-kappa-B on NF-kappa-B through retention in the cytoplasm is exerted primarily through the interaction with RELA. RELA shows a weak DNA-binding site which could contribute directly to DNA binding in the NF-kappa-B complex. Beside its activity as a direct transcriptional activator, it is also able to modulate promoters accessibility to transcription factors and thereby indirectly regulate gene expression (PubMed:29813070). Associates with chromatin at the NF-kappa-B promoter region via association with DDX1. Essential for cytokine gene expression in T-cells. The NF-kappa-B homodimeric RELA-RELA complex appears to be involved in invasin-mediated activation of IL-8 expression. (From Uniprot)
Cys Function & Property
Cys319 is very close to the DNA binding site in space.
- Hydrophobic property:
- SASA:
- Cys319: 53.269 A^2
Protein Sequence
MDDLFPLIFP SEPAQASGPY VEIIEQPKQR GMRFRYKCEG RSAGSIPGER
STDTTKTHPT IKINGYTGPG TVRISLVTKD PPHRPHPHEL VGKDCRDGYY
EADLCPDRSI HSFQNLGIQC VKKRDLEQAI SQRIQTNNNP FHVPIEEQRG
DYDLNAVRLC FQVTVRDPAG RPLLLTPVLS HPIFDNRAPN TAELKICRVN
RNSGSCLGGD EIFLLCDKVQ KEDIEVYFTG PGWEARGSFS QADVHRQVAI
VFRTPPYADP SLQAPVRVSM QLRRPSDREL SEPMEFQYLP DTDDRHRIEE
KRKRTYETFK SIMKKSPFNG PTEPRPPTRR IAVPTRNSTS VPKPAPQPYT
FPASLSTINF DEFSPMLLPS GQISNQALAL APSSAPVLAQ TMVPSSAMVP
LAQPPAPAPV LTPGPPQSLS APVPKSTQAG EGTLSEALLH LQFDADEDLG
ALLGNSTDPG VFTDLASVDN SEFQQLLNQG VSMSHSTAEP MLMEYPEAIT
RLVTGSQRPP DPAPTPLGTS GLPNGLSGDE DFSSIADMDF SALLSQISS
Structural Information
- Known structure with covalent ligand:
- Unknown
- Protein structure:
Related Pathway
- Antifolate resistance
- MAPK signaling pathway
- Ras signaling pathway
- cAMP signaling pathway
- Chemokine signaling pathway
- NF-kappa B signaling pathway
- HIF-1 signaling pathway
- Sphingolipid signaling pathway
- Mitophagy - animal
- PI3K-Akt signaling pathway
- Apoptosis
- Longevity regulating pathway
- Cellular senescence
- Osteoclast differentiation
- Toll-like receptor signaling pathway
- NOD-like receptor signaling pathway
- RIG-I-like receptor signaling pathway
- Cytosolic DNA-sensing pathway
- C-type lectin receptor signaling pathway
- IL-17 signaling pathway
- Th1 and Th2 cell differentiation
- Th17 cell differentiation
- T cell receptor signaling pathway
- B cell receptor signaling pathway
- TNF signaling pathway
- Neurotrophin signaling pathway
- Prolactin signaling pathway
- Adipocytokine signaling pathway
- Relaxin signaling pathway
- Insulin resistance
- Non-alcoholic fatty liver disease (NAFLD)
- AGE-RAGE signaling pathway in diabetic complications
- Cocaine addiction
- Salmonella infection
- Pertussis
- Legionellosis
- Yersinia infection
- Leishmaniasis
- Chagas disease
- Toxoplasmosis
- Amoebiasis
- Tuberculosis
- Hepatitis C
- Hepatitis B
- Measles
- Human cytomegalovirus infection
- Influenza A
- Human papillomavirus infection
- Human T-cell leukemia virus 1 infection
- Kaposi sarcoma-associated herpesvirus infection
- Herpes simplex virus 1 infection
- Epstein-Barr virus infection
- Human immunodeficiency virus 1 infection
- Pathways in cancer
- Transcriptional misregulation in cancer
- Viral carcinogenesis
- Pancreatic cancer
- Prostate cancer
- Chronic myeloid leukemia
- Acute myeloid leukemia
- Small cell lung cancer
- PD-L1 expression and PD-1 checkpoint pathway in cancer
- Inflammatory bowel disease (IBD)
- Fluid shear stress and atherosclerosis
Experimental Evidence
- Cys-directed Mutation
Reference
- Yadav V R, Prasad S, Gupta S C, et al. 3-Formylchromone interacts with cysteine 38 in p65 protein and with cysteine 179 in IκBα kinase, leading to down-regulation of nuclear factor-κB (NF-κB)-regulated gene products and sensitization of tumor cells[J]. Journal of Biological Chemistry, 2012, 287(1): 245-256. 22065587
- Targets
- Mus musculus
- Transcription factor
- Rel homology DNA-binding domain
- Antifolate resistance
- MAPK signaling pathway
- Ras signaling pathway
- CAMP signaling pathway
- Chemokine signaling pathway
- NF-kappa B signaling pathway
- HIF-1 signaling pathway
- Sphingolipid signaling pathway
- Mitophagy - animal
- PI3K-Akt signaling pathway
- Apoptosis
- Longevity regulating pathway
- Cellular senescence
- Osteoclast differentiation
- Toll-like receptor signaling pathway
- NOD-like receptor signaling pathway
- RIG-I-like receptor signaling pathway
- Cytosolic DNA-sensing pathway
- C-type lectin receptor signaling pathway
- IL-17 signaling pathway
- Th1 and Th2 cell differentiation
- Th17 cell differentiation
- T cell receptor signaling pathway
- B cell receptor signaling pathway
- TNF signaling pathway
- Neurotrophin signaling pathway
- Prolactin signaling pathway
- Adipocytokine signaling pathway
- Relaxin signaling pathway
- Insulin resistance
- Non-alcoholic fatty liver disease (NAFLD)
- AGE-RAGE signaling pathway in diabetic complications
- Cocaine addiction
- Salmonella infection
- Pertussis
- Legionellosis
- Yersinia infection
- Leishmaniasis
- Chagas disease
- Toxoplasmosis
- Amoebiasis
- Tuberculosis
- Hepatitis C
- Hepatitis B
- Measles
- Human cytomegalovirus infection
- Influenza A
- Human papillomavirus infection
- Human T-cell leukemia virus 1 infection
- Kaposi sarcoma-associated herpesvirus infection
- Herpes simplex virus 1 infection
- Epstein-Barr virus infection
- Human immunodeficiency virus 1 infection
- Pathways in cancer
- Transcriptional misregulation in cancer
- Viral carcinogenesis
- Pancreatic cancer
- Prostate cancer
- Chronic myeloid leukemia
- Acute myeloid leukemia
- Small cell lung cancer
- PD-L1 expression and PD-1 checkpoint pathway in cancer
- Inflammatory bowel disease (IBD)
- Fluid shear stress and atherosclerosis