N-acylethanolamine-hydrolyzing acid amidase (Mus musculus)
| Basic Information | |
|---|---|
| Short Name | ASAH-like protein, NAAA |
| UNP ID | Q9D7V9 |
| Organism | Mus musculus |
| Cys Site | Cys131 |
| Family/Domain |
Linear amide C-N hydrolases, choloylglycine hydrolase family, Acid ceramidase family |
| Known Ligand | Ligand list |
| Function Type | Metabolic enzyme |
Summary
Protein Function
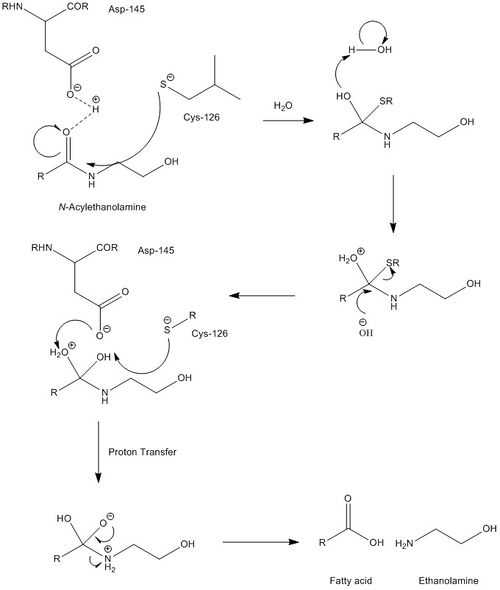
NAAA must operate under acidic conditions (pH ~4.5), and is completely inactivated at a pH of 8. Selective inhibitors of NAAA are ester and amide compounds, such as N-cyclohexanecarbonylpentadecylamine. NAAA is cleaved proteolytically at residue Cys126. NAAA cleaves C-N non-peptide bonds in linear amides, particularly ethanolamides. Its mechanism is quite similar to that of AC, which is further supported by AC's ability to cleave N-acylethanolamines (NAEs), albeit at far lower rates and with different specificities. While mechanistic details are not very well known, catalytic activity of NAAA is thought to be activated by Cys126 and Asp145.
Fatty acid ethanolamines (FAEs) perform several physiological functions, most notably serving as messengers for pain and inflammation. NAAA's are found primarily in the lysosomal compartment of macrophages, in line with most inflammation-related proteins. They perform FAE hydrolysis, the final step in the signaling cascade for pain and inflammation, yielding an ethanolamine and a fatty acid. While it processes the cleavage of many different substrates, NAAA is most active with the substrate N-palmitoylethanolamine, suggesting that this is one of the key messengers of pain. (From Wikipedia)
Cys Function & Property
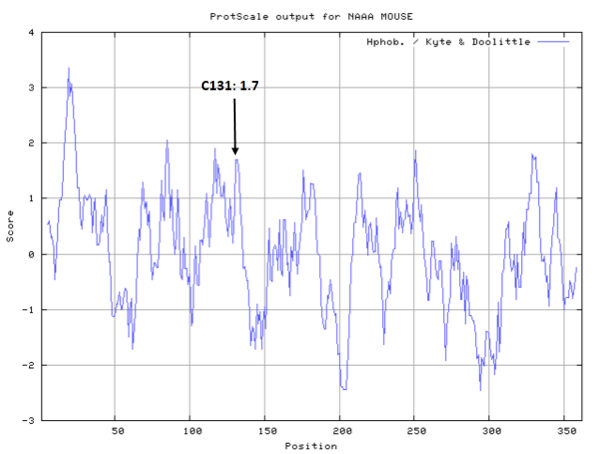
Cys131 is the active site of mouse NAAA.
- Hydrophobic property:
- SASA:
- Cys131: Unknown
Protein Sequence
MGTLATRAAC HGAHLALALL LLLSLSGPWL SAVVPGTPPL FNVSLDAAPE
QRWLPMLRHY DPDFLRTAVA QVIGDRVPQW VLGMVGEIVS KVESFLPQPF
TDEIRSICDS LNLSLADGIL VNLAYEASAF CTSIVAQDSQ GHIYHGRNLD
YPFGKILRKL TANVQFIKNG QIAFTGTTFV GYVGLWTGQS PHKFTISGDE
RDKGWWWENM IAALSLGHSP ISWLIRKTLS ESESFEAAVY TLAKTPLIAD
VYYIVGGTSP KEGVVITRDR GGPADIWPLD PLNGEWFRVE TNYDHWKPAP
KVDDRRTPAI KALNATGQAH LNLETLFQVL SLFPVYNNYT IYTTVMSAAE
PDKYLTMIRN PS
Structural Information
- Known structure with covalent ligand:
- Unknown
- Protein structure:
- Unknown
Related Pathway
- Unknown
Experimental Evidence
- Cys-directed Mutation, Homologous Analysis Of Sequence
Reference
- Armirotti A, Romeo E, Ponzano S, et al. β-Lactones inhibit N-acylethanolamine acid amidase by S-acylation of the catalytic N-terminal cysteine[J]. ACS medicinal chemistry letters, 2012, 3(5): 422-426. 24900487
- Solorzano C, Zhu C, Battista N, et al. Selective N-acylethanolamine-hydrolyzing acid amidase inhibition reveals a key role for endogenous palmitoylethanolamide in inflammation[J]. Proceedings of the National Academy of Sciences, 2009, 106(49): 20966-20971. 19926854