Peroxisome proliferator-activated receptor gamma
| Basic Information | |
|---|---|
| Short Name | PPARγ, PPAR gamma, PPARG |
| UNP ID | P37231 |
| Organism | Homo sapiens |
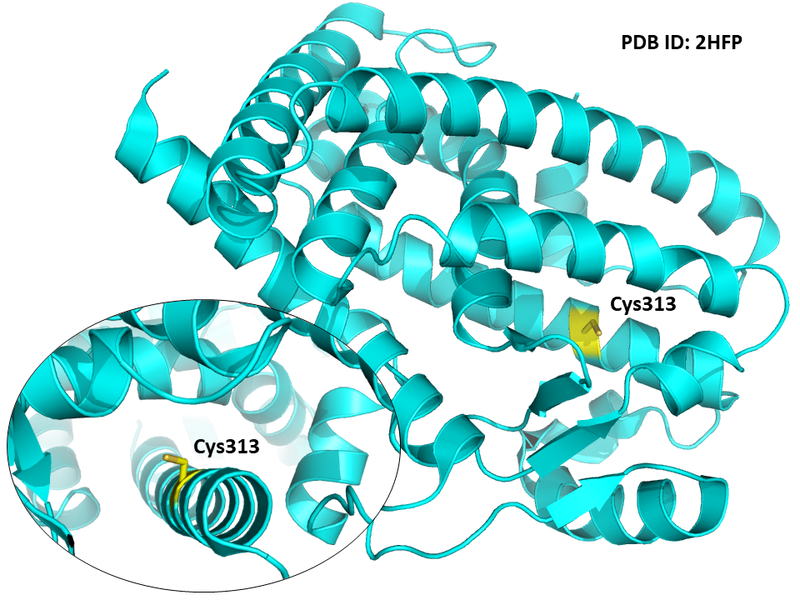
| Cys Site | Cys313 |
| Family/Domain |
Nuclear hormone receptor family, NR1 subfamily |
| Known Ligand | Ligand list |
| Function Type | Metabolic enzyme |
Summary
Protein Function
PPARG regulates fatty acid storage and glucose metabolism. The genes activated by PPARG stimulate lipid uptake and adipogenesis by fat cells. PPARG knockout mice fail to generate adipose tissue when fed a high-fat diet.
This gene encodes a member of the peroxisome proliferator-activated receptor (PPAR) subfamily of nuclear receptors. PPARs form heterodimers with retinoid X receptors (RXRs) and these heterodimers regulate transcription of various genes. Three subtypes of PPARs are known: PPAR-alpha, PPAR-delta, and PPAR-gamma. The protein encoded by this gene is PPAR-gamma and is a regulator of adipocyte differentiation. Alternatively spliced transcript variants that encode different isoforms have been described.
Many naturally occurring agents directly bind with and activate PPAR gamma. These agents include various polyunsaturated fatty acids like arachidonic acid and arachidonic acid metabolites such as certain members of the 5-Hydroxyicosatetraenoic acid and 5-oxo-eicosatetraenoic acid family, e.g. 5-oxo-15(S)-HETE and 5-oxo-ETE or 15-Hydroxyicosatetraenoic acid family including 15(S)-HETE, 15(R)-HETE, and 15(S)-HpETE. The phytocannabinoid tetrahydrocannabinol (THC), its metabolite THC-COOH, and its synthetic analog ajulemic acid (AJA). The activation of PPAR gamma by these and other ligands may be responsible for inhibiting the growth of cultured human breast, gastric, lung, prostate and other cancer cell lines. (From Wikipeida)
Cys Function & Property
Several endogenous fatty acid metabolites have been identified as PPARγ ligands. These fatty acid compounds could covalently bind to the PPARγ ligand binding domain (LBD) via a unique cysteine, which is reportedly critical for receptor activation. (PMID: 18977231)
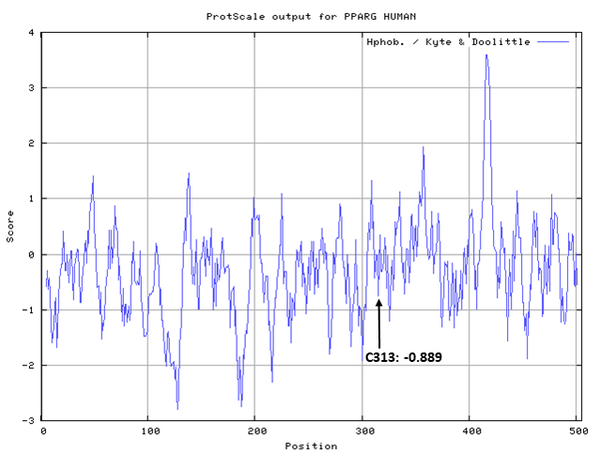
- Hydrophobic property:
- SASA:
- Cys313: 40.483 A^2
Protein Sequence
MGETLGDSPI DPESDSFTDT LSANISQEMT MVDTEMPFWP TNFGISSVDL
SVMEDHSHSF DIKPFTTVDF SSISTPHYED IPFTRTDPVV ADYKYDLKLQ
EYQSAIKVEP ASPPYYSEKT QLYNKPHEEP SNSLMAIECR VCGDKASGFH
YGVHACEGCK GFFRRTIRLK LIYDRCDLNC RIHKKSRNKC QYCRFQKCLA
VGMSHNAIRF GRMPQAEKEK LLAEISSDID QLNPESADLR ALAKHLYDSY
IKSFPLTKAK ARAILTGKTT DKSPFVIYDM NSLMMGEDKI KFKHITPLQE
QSKEVAIRIF QGCQFRSVEA VQEITEYAKS IPGFVNLDLN DQVTLLKYGV
HEIIYTMLAS LMNKDGVLIS EGQGFMTREF LKSLRKPFGD FMEPKFEFAV
KFNALELDDS DLAIFIAVII LSGDRPGLLN VKPIEDIQDN LLQALELQLK
LNHPESSQLF AKLLQKMTDL RQIVTEHVQL LQVIKKTETD MSLHPLLQEI
YKDLY
Structural Information
- Known structure with covalent ligand:
- Protein structure:
Related Pathway
- PPAR signaling pathway
- AMPK signaling pathway
- Longevity regulating pathway
- Osteoclast differentiation
- Thermogenesis
- Huntington disease
- Pathways in cancer
- Transcriptional misregulation in cancer
- Thyroid cancer
Experimental Evidence
- Crystallography, Isotope Labeling
Reference
- Waku T, Shiraki T, Oyama T, et al. Structural insight into PPARγ activation through covalent modification with endogenous fatty acids[J]. Journal of molecular biology, 2009, 385(1): 188-199. 18977231
- Shiraki T, Kamiya N, Shiki S, et al. α, β-unsaturated ketone is a core moiety of natural ligands for covalent binding to peroxisome proliferator-activated receptor γ[J]. Journal of Biological Chemistry, 2005, 280(14): 14145-14153. 15695504
- Hiromori Y, Nishikawa J, Yoshida I, et al. Structure-dependent activation of peroxisome proliferator-activated receptor (PPAR) γ by organotin compounds[J]. Chemico-biological interactions, 2009, 180(2): 238-244. 19497422
- Waku T, Shiraki T, Oyama T, et al. The nuclear receptor PPARγ individually responds to serotonin‐and fatty acid‐metabolites[J]. The EMBO journal, 2010, 29(19): 3395-3407. 20717101