Potassium-transporting ATPase alpha chain 1 (Sus scrofa)
| Basic Information | |
|---|---|
| Short Name | ATP4A |
| UNP ID | P19156 |
| Organism | Sus scrofa |
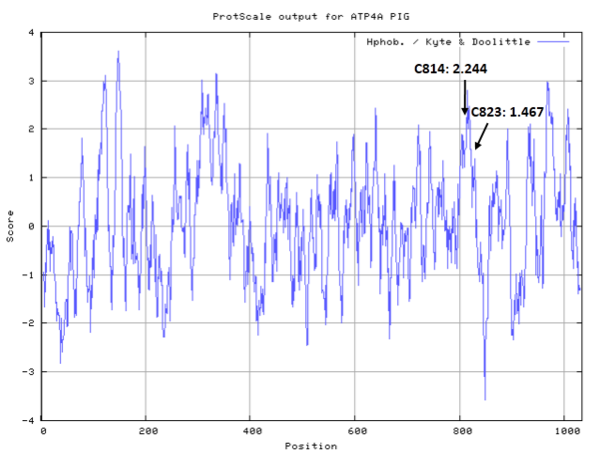
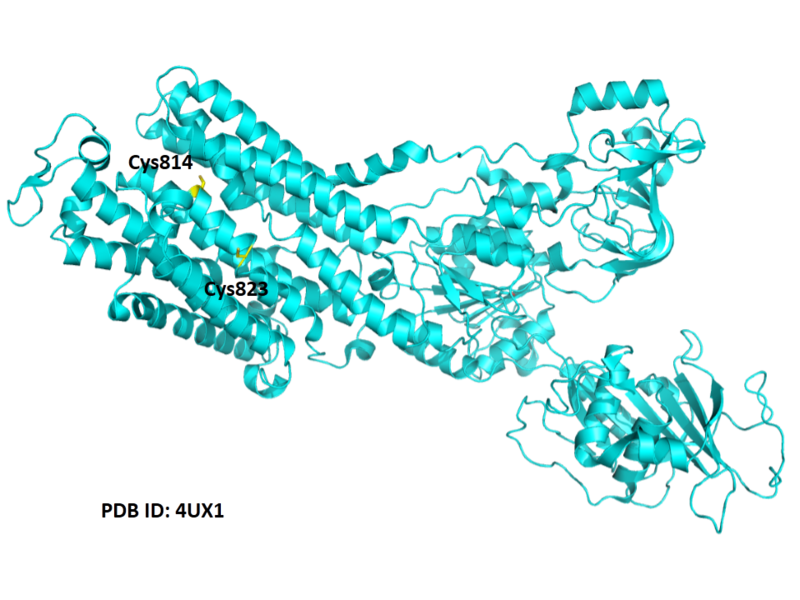
| Cys Site | Cys814, Cys823 |
| Family/Domain |
Cation transporting ATPase, C-terminus, Cation transport ATPase (P-type) family, Type IIC subfamily |
| Known Ligand | Ligand list |
| Function Type | Ion channel |
Summary
Protein Function
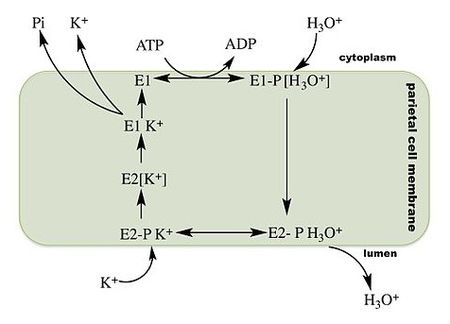
The gastric hydrogen potassium ATPase or H+/K+ ATPase is the proton pump of the stomach. It exchanges potassium from the intestinal lumen with cytoplasmic hydronium and is the enzyme primarily responsible for the acidification of the stomach contents and the activation of the digestive enzyme pepsin.
H+/K+ ATPase is a P2-type ATPase, a member of the eukaryotic class of P-type ATPases. Like the Ca2+ and the Na+/K+ ATPases, the H+/K+ ATPase functions as an α, β protomer. Unlike other eukaryotic ATPases, the H+/K+ ATPase is electroneutral, transporting one proton into the stomach lumen per potassium retrieved from the gastric lumen. As an ion pump the H+/K+ ATPase is able to transport ions against a concentration gradient using energy derived from the hydrolysis of ATP. Like all P-type ATPases, a phosphate group is transferred from adenosine triphosphate (ATP) to the H+/K+ ATPase during the transport cycle. This phosphate transfer powers a conformational change in the enzyme that helps drive ion transport. (From Wikipedia)
Cys Function & Property
- Hydrophobic property:
- SASA:
- Cys814: 24.44 A^2
- Cys823: 27.265 A^2
Protein Sequence
MGKAENYELY QVELGPGPSG DMAAKMSKKK AGRGGGKRKE KLENMKKEME
INDHQLSVAE LEQKYQTSAT KGLSASLAAE LLLRDGPNAL RPPRGTPEYV
KFARQLAGGL QCLMWVAAAI CLIAFAIQAS EGDLTTDDNL YLALALIAVV
VVTGCFGYYQ EFKSTNIIAS FKNLVPQQAT VIRDGDKFQI NADQLVVGDL
VEMKGGDRVP ADIRILQAQG RKVDNSSLTG ESEPQTRSPE CTHESPLETR
NIAFFSTMCL EGTAQGLVVN TGDRTIIGRI ASLASGVENE KTPIAIEIEH
FVDIIAGLAI LFGATFFIVA MCIGYTFLRA MVFFMAIVVA YVPEGLLATV
TVCLSLTAKR LASKNCVVKN LEAVETLGST SVICSDKTGT LTQNRMTVSH
LWFDNHIHSA DTTEDQSGQT FDQSSETWRA LCRVLTLCNR AAFKSGQDAV
PVPKRIVIGD ASETALLKFS ELTLGNAMGY RERFPKVCEI PFNSTNKFQL
SIHTLEDPRD PRHVLVMKGA PERVLERCSS ILIKGQELPL DEQWREAFQT
AYLSLGGLGE RVLGFCQLYL SEKDYPPGYA FDVEAMNFPT SGLSFAGLVS
MIDPPRATVP DAVLKCRTAG IRVIMVTGDH PITAKAIAAS VGIISEGSET
VEDIAARLRV PVDQVNRKDA RACVINGMQL KDMDPSELVE ALRTHPEMVF
ARTSPQQKLV IVESCQRLGA IVAVTGDGVN DSPALKKADI GVAMGIAGSD
AAKNAADMIL LDDNFASIVT GVEQGRLIFD NLKKSIAYTL TKNIPELTPY
LIYITVSVPL PLGCITILFI ELCTDIFPSV SLAYEKAESD IMHLRPRNPK
RDRLVNEPLA AYSYFQIGAI QSFAGFTDYF TAMAQEGWFP LLCVGLRPQW
ENHHLQDLQD SYGQEWTFGQ RLYQQYTCYT VFFISIEMCQ IADVLIRKTR
RLSAFQQGFF RNRILVIAIV FQVCIGCFLC YCPGMPNIFN FMPIRFQWWL
VPMPFGLLIF VYDEIRKLGV RCCPGSWWDQ ELYY
Structural Information
- Known structures with covalent ligands:
- Unknown
- Protein structure:
Related Pathway
Experimental Evidence
- Tryptic Digest, Isotope Labeling, Tricine-SDS-PAGE, Homologous Analysis of Sequence
Reference
- Shin J M, Besancon M, Simon A, et al. The site of action of pantoprazole in the gastric H+/K+-ATPase[J]. Biochimica et Biophysica Acta (BBA)-Biomembranes, 1993, 1148(2): 223-233. 8389196
- Morii M, Takata H, Fujisaki H, et al. The potency of substituted benzimidazoles such as E3810, omeprazole, Ro 18-5364 to inhibit gastric H+, K+-ATPase is correlated with the rate of acid-activation of the inhibitor[J]. Biochemical pharmacology, 1990, 39(4): 661-667. 2154989