Protein-arginine deiminase type-2 (Homo sapiens)
| Basic Information | |
|---|---|
| Short Name | PADI2, PDI2 |
| UNP ID | Q9ULC6 |
| Organism | Homo sapiens |
| Cys Site | Cys647 |
| Family/Domain | Protein arginine deiminase family |
| Known Ligand | Ligand list |
| Function Type | Metabolic enzyme |
Summary
Protein Function
Protein-arginine deiminase type-2 is a member of the peptidyl arginine deiminase family of enzymes, which catalyze the post-translational deimination of proteins by converting arginine residues into citrullines in the presence of calcium ions. The family members have distinct substrate specificities and tissue-specific expression patterns. The type II enzyme is the most widely expressed family member. Known substrates for this enzyme include myelin basic protein in the central nervous system and vimentin in skeletal muscle and macrophages. This enzyme is thought to play a role in the onset and progression of neurodegenerative human disorders, including Alzheimer disease and multiple sclerosis, and it has also been implicated in glaucoma pathogenesis. This gene exists in a cluster with four other paralogous genes. (From Wikipedia)
Cys Function & Property
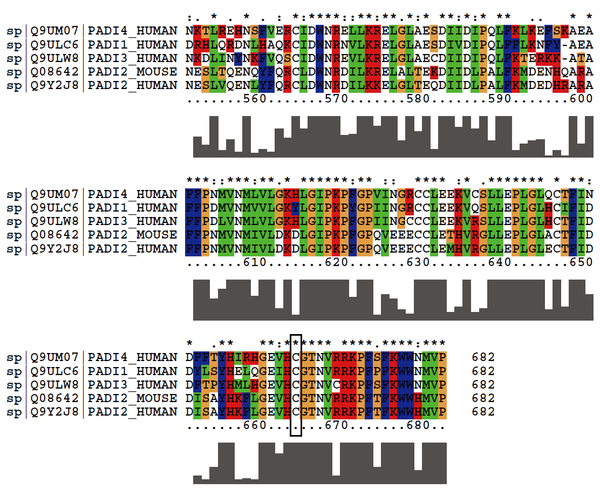
Cys647 is the active site of human PADI2. And this site is quite conserved in PADIs.
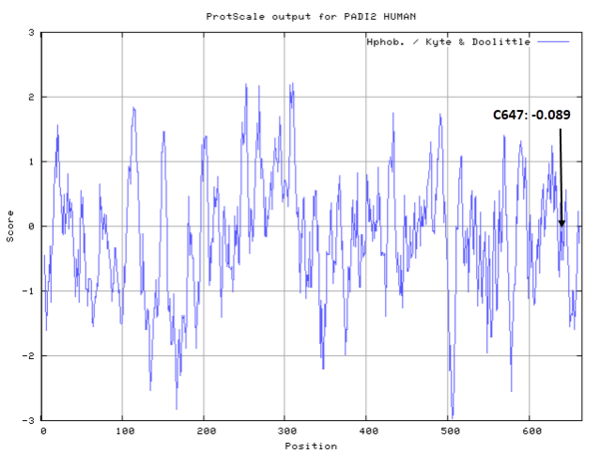
- Hydrophobic property:
- SASA:
- Cys647: 6.833 A^2
Protein Sequence
MLRERTVRLQ YGSRVEAVYV LGTYLWTDVY SAAPAGAQTF SLKHSEHVWV
EVVRDGEAEE VATNGKQRWL LSPSTTLRVT MSQASTEASS DKVTVNYYDE
EGSIPIDQAG LFLTAIEISL DVDADRDGVV EKNNPKKASW TWGPEGQGAI
LLVNCDRETP WLPKEDCRDE KVYSKEDLKD MSQMILRTKG PDRLPAGYEI
VLYISMSDSD KVGVFYVENP FFGQRYIHIL GRRKLYHVVK YTGGSAELLF
FVEGLCFPDE GFSGLVSIHV SLLEYMAQDI PLTPIFTDTV IFRIAPWIMT
PNILPPVSVF VCCMKDNYLF LKEVKNLVEK TNCELKVCFQ YLNRGDRWIQ
DEIEFGYIEA PHKGFPVVLD SPRDGNLKDF PVKELLGPDF GYVTREPLFE
SVTSLDSFGN LEVSPPVTVN GKTYPLGRIL IGSSFPLSGG RRMTKVVRDF
LKAQQVQAPV ELYSDWLTVG HVDEFMSFVP IPGTKKFLLL MASTSACYKL
FREKQKDGHG EAIMFKGLGG MSSKRITINK ILSNESLVQE NLYFQRCLDW
NRDILKKELG LTEQDIIDLP ALFKMDEDHR ARAFFPNMVN MIVLDKDLGI
PKPFGPQVEE ECCLEMHVRG LLEPLGLECT FIDDISAYHK FLGEVHCGTN
VRRKPFTFKW WHMVP
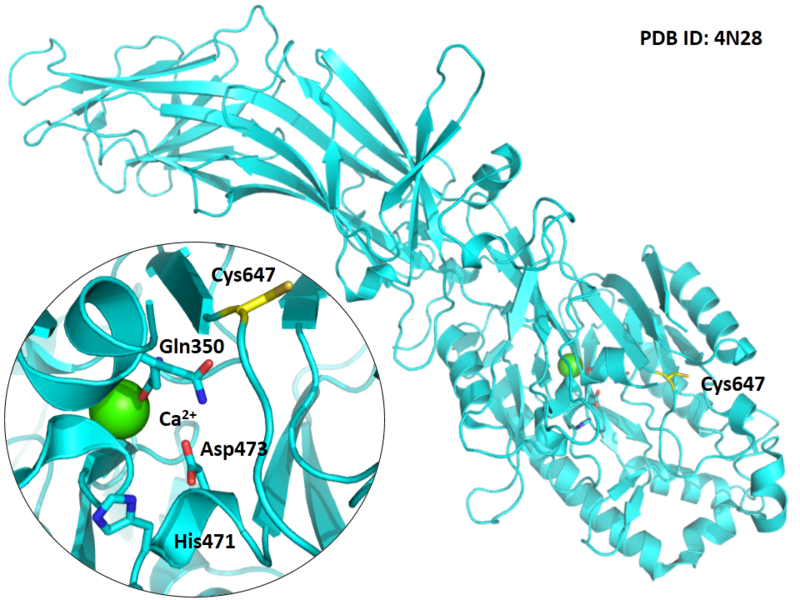
Structural Information
- Known structure with covalent ligand:
- Unknown
- Protein structure:
Related Pathway
- Unknown
Experimental Evidence
- Homologous Analysis of Sequence, Mass Spectrometry
Reference
- Jones J E, Slack J L, Fang P, et al. Synthesis and screening of a haloacetamidine containing library to identify PAD4 selective inhibitors[J]. ACS chemical biology, 2011, 7(1): 160-165. 22004374